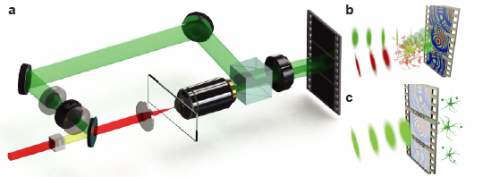
Schematics of the SHG holographic microscope. (A) Off-axis holography setup is configured as a modified Mach–Zehnder interferometer. A detailed description is provided in Methods. (B) Representation of a sample emitting SHG, which is combined with the reference on a high-speed camera. (C) Holograms are numerically reconstructed, forming video-rate 3D information from a 2D hologram image. Copyright © PNAS, doi:10.1073/pnas.1306856110
(Phys.org) —In the world of biomedical science, optical microscopy rules – and nonlinear optical microscopy, which uses ultrashort pulse lasers as the illumination source, allows researchers to glean much greater detail from biological specimens. That being said, the technique's weak signal levels significantly limit 3D image acquisition rates. Recently, however, scientists at Colorado State University employed 3D second harmonic generation achieved frame rates over 8,000 times faster than is possible with current nonlinear optical microscopy. (In second harmonic generation, or SHG, photons interact with a nonlinear material to form new photons with twice the energy and, therefore, twice the frequency and half the wavelength. While conventional optical microscopes obtain contrast by detecting variations in optical density, path length, or refractive index of the specimen, a second harmonic imaging microscope derives contrast from variations in a specimen's ability to generate second harmonic light from incident laser light.) Moreover, the scientists introduced new methods that greatly improve the ability to quantify signal-to-noise quality. The researchers state that their study allows nonlinear optical imaging to study behavior that current experimental methodologies are unable to capture, such as neural circuit dynamics.
Prof. Randy A. Bartels discussed the paper that he, David R. Smith and David G. Winters published recently in Proceedings of the National Academy of Sciences. "Our current configuration is capable of imaging continuously at essentially 10 mm/s for bright enough objects," Bartels tells Phys.org. "In general, the main challenge in achieving this is making sure that the fringes don't wash out for a given velocity – and we provide theory to easily calculate these conditions, such that the configuration in this paper was capable of recording holograms of objects moving at this velocity. If we consider an object moving at 10 mm/s," he explains, "then in a 1-ms integration time (which we can easily do continuously), we need to resolve an object displacement of ~ 10 micron, which is roughly twenty times our current imaging resolution." Although faster imaging is possible for a bright enough object with the camera plane moved farther away from the image plane, he adds, the object scatter enough second harmonic light to form a good hologram – and while for any particular object this is limited by the hyperpolarizability of constituent harmonophores (molecules or particles that produce second harmonic generation), the researchers can increase brightness with shorter illumination pulses.
"We're already using SHG to observe endogenous harmonophores," Bartels continues, and are now studying live dynamics of a number of processes such as muscle contractions in embryonic models, and dynamics of structural tissues such as tendons, under high strain rates." The team is also taking beginning steps to image action potentials of neurons.
3D SHG video recording of a potato starch flow recorded at 1,594.6 fps. (Scale bar: 10 μm.) Copyright © PNAS, doi:10.1073/pnas.1306856110
The main challenges in observing dynamics in live tissue, Bartels says, are the same as needed to speed up imaging to 10 mm/s – namely, brighter SHG scattering (which for a given specimen can only be increased by decreasing the illumination pulse duration) and optimizing the detection geometry. "While in this study we used a bulk ytterbium (Yb) solid state laser built in my laboratory that makes 350 femtoseconds laser pulses, we're rebuilding the microscope with a new laser source that generates 80 fs pulses. This should increase our signals and top-end image field velocity by more than a factor of four. Also, it's important to move the camera plane as close to the object as possible," he points out, "as long as the fringes aren't degraded by the moving objects." The current paper provides a framework for evaluating an optimal experimental setup.
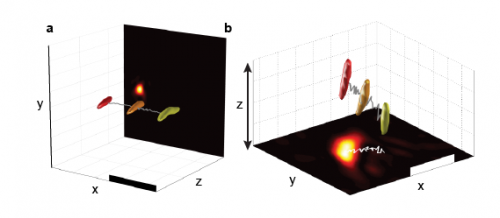
High-speed 4D reconstructions with centroid tracking. (A) 3D reconstructions of potato starch granules with a trace marking the centroid path. Yellow indicates t = 0 ms, orange indicates t = 32 ms, and red indicates t = 63 ms. (B) 3D volume reconstructions show movement in three dimensions. Yellow indicates t = 0 ms, orange indicates t = 50 ms, and red indicates t = 92 ms. (Scale bars: 10 μm.) (A video recording of the moving particles is provided in Movie S1.) Copyright © PNAS, doi:10.1073/pnas.1306856110
The scientists have also been able to exploit coherent scattering of second harmonic light from an entire specimen volume. "We've already observed the benefits of coherent volume scattering by comparing images captured with SHG holography and more conventional laser scanning second harmonic holography," Bartels notes. "In structures, such as sarcomere segments in muscle fibers, we observed significantly brighter SHG signals from holography as compared to laser scanning SHG microscopy." He adds that the differences were less pronounced for more compact objects.
Given these challenges, Bartels says that increasing imaging speed required us to understand how to optimize the signal to noise levels of the SHG hologram and balance that optimization with an experimental design that prevented holographic fringes from washing out during high speed object motion. "We developed rigorous metrics for independently estimating noise and signal levels of a hologram, and optimized image speed, sensitivity, and signal to noise levels to obtain the optimal performance from the experimental system." Their results suggested improvements that they plan to implement in their next-generation system.
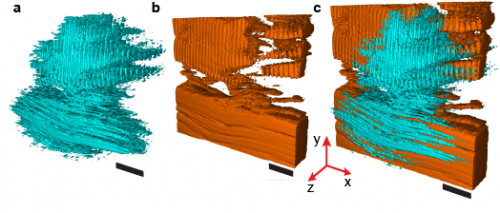
Comparison of SHG holographic reconstructions with LSM 3D images. All images were taken from a 50-μm thick slice of mouse skeletal muscle. (A) Holographic reconstruction shows connective tissue and sarcomere structures. (B) LSM 3D image of the same tissue region. (C) Volumetric overlay of the 3D images in A and B. (Scale bars: 10 μm.) Another volume reconstruction comparison is provided in SI Methods, section 13, Fig. S7. Copyright © PNAS, doi:10.1073/pnas.1306856110
Bartels tells Phys.org that while laser scanning is the standard approach to imaging with nonlinear optical contrast, since the beams must be scanned 3D image update rates are limited – and the capture of fast dynamics are restricted to 1D or 2D images, or charting a trajectory through the specimen. "A major problem is that live specimen motion provides changes to the image that distort measurements and make it impossible to track high speed behaviors. On the other hand, SHG holography is a non-scanning technique that captures 3D information in a single shot for each volume image frame."
Bartels adds that in addition to neural circuit dynamics, the scientists are interested in observing the high speed dynamics of muscle contraction and connective tissues subjected to high speed stresses, and observing the resultant strains. "Current imaging techniques lack the ability to observe the effect of such trauma on tissues, as well as to follow the subsequent strains and relaxation or damage that occurs. SHG holography will be able to capture a time sequence of images from these tissues to better understand tissue properties. Moreover," he continues, "SHG holography opens up imaging of dynamics that were previously not observable. As biological functions are determined by 3D organization in tissues, the ability to study high speed dynamics of biological functions – or the response of biological systems and tissues to rapid external stimuli, perturbations, or trauma – will expand our understanding of the biology, and may inform our ability to treat injuries."
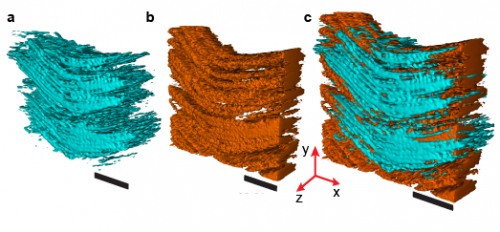
Comparison of SHG holographic reconstructions with LSM 3D images. All images were taken from a 50-μm thick slice of mouse skeletal muscle. (A) Holographic reconstruction of collagen-rich connective tissue found in the skeletal muscle slice. (B) LSM 3D image of the same tissue region. (C) Volumetric overlay of the 3D images in A and B. (Scale bars: 10 μm.) Copyright © PNAS, doi:10.1073/pnas.1306856110
Moving forward, says Bartels, the team is working to develop and integrate other methods of high-speed 3D imaging into their SHG holography platform – specifically, 3D fluorescent imaging capabilities, which is more challenging because fluorescent light lacks the coherence that we are able to exploit for high speed SHG 3D imaging. "We're primarily interested in studying fast dynamics in live biological tissues and systems, and are working with a number of collaborators to look at various systems that display native SHG emission. We're also expanding the capabilities of the microscope to increase axial resolution through new hologram reconstruction algorithms and modifications of the microscope design." Their goal is to study dynamics that have been too fast to be captured by current microscopy tools and better understand that relevant biological processes.
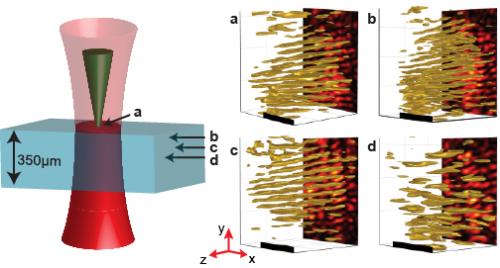
3D reconstructions of 350-μm thick mouse skeletal muscle at depths up to 150 μm. With the focus on the surface of the tissue as indicated in the cartoon, an SHG hologram is recorded (A) and reconstructed 75 μm deep into the tissue (B), 100 μm deep into the tissue (C), and 150 μm deep into the tissue (D). Holograms were recorded with an exposure time of 1 ms (A–C) or 4 ms (D). (Scale bars: 10 μm.) Copyright © PNAS, doi:10.1073/pnas.1306856110
"Beyond biological applications," Bartels concludes, "I can imagine high throughput imaging of starch content for studying or monitoring biofuel production. Alternatively, kinetics of crystal formation and growth for some classes of crystals, including protein crystals, could be monitored."
More information: Submillisecond second harmonic holographic imaging of biological specimens in three dimensions, PNAS November 12, 2013 vol. 110 no. 46 18391-18396, doi:10.1073/pnas.1306856110
Journal information: Proceedings of the National Academy of Sciences
© 2013 Phys.org. All rights reserved.