Combining chromatography, proteomics and database searching identifies hard-to-find heme proteins
(Phys.org) —Iron is a critical part of many biological processes; however, it is often not biologically available or it can be toxic in high quantities. So, biological systems have developed intricate methods for its use and storage. Scientists at DOE's Pacific Northwest National Laboratory combined chromatography, proteomics and a database search strategy to find higher numbers of modified iron-containing protein fragments (called peptides) that play an important role in respiration, metal reduction and nitrogen fixation by environmental microbes. The microbes containing these proteins are being studied because of their potential use in microbial fuel cells and electrosynthesis of valuable biomaterials.
The modifying group of atoms, called heme c, is an important iron-containing post-translational modification found in many proteins. Until recently, it was hard to find. The PNNL scientists combined a heme c protein affinity purification strategy called histidine affinity chromatography (HAC) with enhanced database searching. This combination confidently identified heme c peptides in liquid chromatography-tandem mass spectrometry (LC-MS/MS) experiments by as much as 100-fold in some cases. They used proteomics capabilities housed at DOE's EMSL, located at PNNL.
"Several proteomics studies have analyzed the expression of c-type cytochromes under various conditions," said PNNL researcher Dr. Eric Merkley, lead author of a paper that appeared in the Journal of Proteome Research. "A shared feature of these studies is that the cytochrome-rich fractions, the cell envelope or extracellular polymeric substance, were purified and explicitly analyzed to efficiently detect cytochromes. Analyses of large-scale proteomics datasets have typically suggested that c-type cytochromes, particularly the heme c peptides, are under-represented."
Many environmentally important microbes and microbial communities are rich in c-type cytochromes. Combining HAC and data analysis tailored to the unique properties of heme c peptides should enable more detailed study of the role of c-type cytochromes in these microbes and microbial communities.
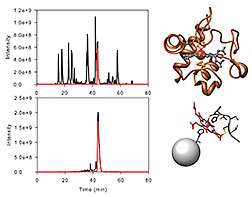
The scientists adapted HAC to enrich heme c protein fragments, or peptides, from purified bovine heart cytochrome c, decaheme cytochromes from Sideroxydans lithotrophicus ES-1 and Shewanella oneidensis MR-1, and from mixtures of Escherichia coli peptides with a standard heme peptide at varying ratios. LC-MS/MS analysis of the samples yielded significantly more matches to heme c peptides than in samples not enriched by HAC.
They also developed a proteomics database search strategy that accounts for the unique physicochemical properties of heme c peptides. Combining affinity chromatography and heme-specific informatics yielded 20- to 100-fold increases in the number of peptide-spectrum matches for bovine cytochrome c.
The scientists are applying and evaluating the methodology for whole-proteome analysis of metal-reducing bacteria. Supplemented by additional fractionation steps, the method could be applied to more complex samples. In addition, HAC combined with mass spectrometric or other methods could become a useful tool for studying the biology of c-type cytochrome biogenesis.
Journal information: Journal of Proteome Research
Provided by US Department of Energy