September 3, 2012 feature
Focusing the phenotype: Controlling genetic expression through external feedback
(Phys.org)—Gene expression plays a central role in the orchestration of virtually all cellular processes. While inducible promoters have proven invaluable in understanding regulatory networks by modifying gene expression levels, their use has faced some shortcomings. Specifically, their utility is constrained to research studying relatively short-term, population-scale effects. Recently, however, scientists at Institut National de Recherche en Informatique et Automatique (INRIA) and at Centre National de la Recherche Scientifique (CNRS), France, have demonstrated that implementing an external feedback loop allows single-cell gene expression to be accurately and tightly controlled over many generations. The team accomplished this by developing a real-time, closed-loop control gene expression platform integrating microscopy, microfluidics, and original software for automated imaging, quantification, and model predictive control. They state that their study demonstrates long-term control with both time-constant and time-varying target expression profiles, at the population and single-cell levels, shows that real-time control can limit the effects of gene expression stochasticity, and anticipate that their method will be useful for improving the performance of complex, synthetically engineered cellular networks.
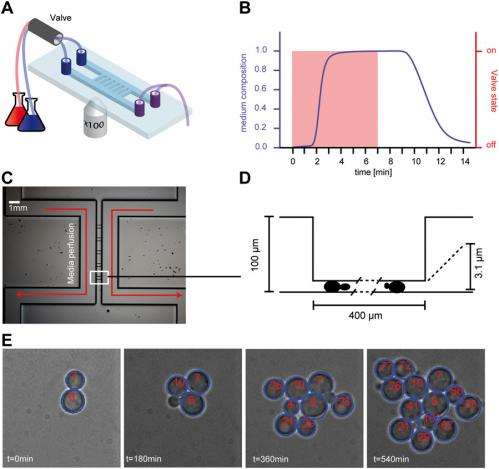
Research Scientist Gregory Batt describes the challenges the research team faced in this study in designing an external feedback loop and platform to tightly control gene expression over many cell generations. "Because gene expression is a relatively slow process," Batt tells Phys.org, "we had to do long-term experiments – each lasting 15 hours – to demonstrate the effective control of gene expression. Each experiment corresponded to many yeast cell generations, and every part of the platform had to be adapted accordingly."
For example, Batt continues, with the first microfluidic device they used, cells were directly under the flow of nutrient media. This allowed them to change the cells' environment very rapidly, but when the cells divided, they were detaching from the bottom glass slide where they were "glued" and were then washed away. "We therefore had to adapt our device by making a type of trap where cells were protected from the main media flow."
In terms of original software for automated imaging, quantification, and model predictive control, Batt adds, the quantification of the protein concentration via the cell fluorescence measurements required particular attention. "Robustness was the issue here. Because we did real-time control, control decisions were made online. Therefore, all the steps of image analysis had to be done in a completely automated manner." They therefore made a significant effort in developing robust routines for autofocus (to maintain good image quality over time), image segmentation (to detect the contours of all cells in the image), and cell tracking (to follow individual cells over time and detect newborn cells). "In particular," notes Batt, "the imaging frequency had to be carefully chosen. Too many images are noxious for cells due to phototoxicity and photobleaching. On the other hand, too few images degrade the performance of autofocus and cell tracking."
In addressing these challenges, says Batt, "our key intuition was probably that automated control techniques and modeling will be effective enough to obtain quantitatively good performance despite the technological issues mentioned above – and even more importantly, despite the significant inertia and stochasticity of gene expression processes. Indeed, there is a significant lag between the initiation of the transcription on the chromosome and the effective increase of cell fluorescence. Transcription, translation, and protein folding take time."
Therefore, Batt explains, the influences of recent past events are not apparent and must be taken into account via modeling. "Unfortunately, modeling is not trivial since gene expression is also a stochastic process. Confronted with the same stimulation, two cells do not respond identically. Even more problematic is the fact that the same cell confronted twice with the same stimulation does not respond twice the same." Therefore, notes Batt, the main open question at the beginning of the project was whether good performances could be obtained via model predictive control despite the significant inertia and stochasticity of gene expression processes.
Batt describes other innovations that might be developed and applied to the current experimental design. "With our setting, we trigger gene expression by applying osmotic shocks – that is, by changing the normal cell environment by a salt-rich or sugar-rich environment, and by highjacking the normal cell hyperosmotic response so that cells produce fluorescent proteins when shocked. Therefore, by design, all cells feel the same stimulation. Recently-developed novel optogenetic techniques allow individual stimulation of cells in a population: cellular processes can be activated or inactivated simply by exposing a cell to light. "The use of optogenetic techniques within our control platform should expand our control capabilities: each cell within a population could be controlled individually to fulfill the population objective."
To probe the functioning of dynamical systems, scientists generally observe how the system reacts following some perturbations – an approach that's standard for studying the functioning of cellular processes. "However, current methods for perturbing protein levels – most notably the use of inducible promoters and RNA interference – are fairly limited in terms of duration and quantitative accuracy," Batt points out. "Moreover, it is not possible to obtain well-controlled time-varying profiles of protein concentrations. Our platform for real-time control of gene expression has been developed precisely to address these limitations. "
Synthetic biology aims at reprogramming cells so that they implement useful functions, and currently its typical target applications include biofuel production, bioremediation and even cellular therapies. In practice, bioengineers take advantage of the metabolic and information processing capabilities of cells to reengineer cells towards a desired objective. "However," Batt comments, "it's often nontrivial to implement robustly all functions using exclusively biological building blocks. By offering the possibility to use an external control loop, our project offers the possibility to develop mixed systems in which some elements are implemented in vivo using synthetic biology principles and some elements are implemented in silico using standard programming principles."
An important result of the research is that the findings provide evidence that real-time control can dynamically limit the effects of gene expression stochasticity. "We performed two types of experiments," Batt notes, "In the first, we measured the fluorescence of all the cells in the microscope field of view, and used the mean cell fluorescence as a control criterion. In the second case, we only focus on one cell, randomly chosen at the beginning of the experiment, and defined the fluorescence of this cell as the control criterion. Then, we observed that the fluctuations around the target value of the fluorescence of a cell were statistically smaller when cells were individually controlled than when they were controlled as a member of a population."
In addition to systems and synthetic biology applications, Batt concludes, "Our platform can also be of interest to computational biologists, since it paves the way for online model selection and online model tuning." In this setting, experiments are carried out, analyzed and interpreted in real-time, so that models can be challenged in the most efficient manner.
More information: Long-term model predictive control of gene expression at the population and single-cell levels, PNAS August 28, 2012 vol. 109 no. 35 14271-14276, doi:10.1073/pnas.1206810109
Journal information: Proceedings of the National Academy of Sciences
Copyright 2012 Phys.org
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.