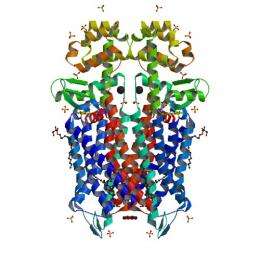
Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Image courtesy of Aashish Manglik, Andrew C. Kruse, Tong Sun Kobilka, Foon Sun Thian, Jesper M. Mathiesen, Roger K. Sunahara, Leonardo Pardo, William I. Weis, Brian K. Kobilka & Sébastien Granier
(Phys.org) -- Researchers and doctors have gleaned new clues to the molecular mechanisms behind some of the most addictive substances in the world, thanks to two new studies that uncovered the structures of some of the most intricate and challenging proteins ever analyzed on the atomic level.
In separate studies recently reported in Nature (1, 2), users of the Advanced Photon Source (APS) at the U.S. Department of Energy’s (DOE) Argonne National Laboratory determined the composition of brain receptors that bind to opioids, the class of molecules that includes morphine, heroin and oxycodone.
The experiments show that the binding sites of both µ (mu) and ĸ (kappa) opioid receptors are relatively large and open, which researchers believe could help explain both why they recognize an array of molecules and why opioids are processed so quickly by the brain.
Opioid receptors belong to a class of molecules known as G-protein coupled receptors (GPCRs), which are typically proteins that sense other molecules that exist in the area immediately outside the cell membrane. For decades, the exact configurations of GPCRs have flummoxed scientists.
“The GPCRs are such a diverse family in terms of their function, but until very recently we just didn’t have the technology required to learn about their structures,” said Argonne senior scientist Robert Fischetti.
When a GPCR senses its target molecule, it triggers a series of changes within the cell. GPCRs help to fulfill a number of different biological roles, including enabling vision and the sense of smell, and – in the case of receptors for opioids and other brain chemicals – regulating behavior and mood.
According to Insight Pharma Reports, roughly 30 to 40 percent of all current drug development is targeted for GPCRs.
At the APS, Fischetti and Janet Smith with the University of Michigan manage a suite of National Institutes of Health-supported beamlines where researchers use high-energy X-rays to probe the molecular structures of many different proteins. These beamlines, named for the National Institute of General Medicine Sciences and National Cancer Institute Collaborative Access Team (GM/CA-CAT), allow visiting protein crystallographers to work around the clock to study proteins involved in biological pathways that regulate both normal functioning and disease. These beamlines specializes in intense, tunable micro-beams for crystallography.
“Argonne leads the world when it comes to developing and providing access to new crystallographic instruments and techniques,” said Brian Kobilka, a crystallographer from Stanford University who headed one of the two studies.
According to Argonne crystallographer Ruslan (Nukri) Sanishvili, one principal feature of GM/CA-CAT that makes it especially attractive for protein crystallography is the development of a device known as a quad collimator, which allows researchers to shrink the size of the X-ray beam with a click of a button. This “minibeam” device -- together with a special “rastering” software program -- enables the analysis of significantly smaller or otherwise inferior crystals than ever before, Sanishvili said.
“GPCRs are known for being relatively difficult to crystallize – that was one of the main reasons the biological community had been having such a hard time understanding their structures,” said Michael Becker, another Argonne crystallographer. “The minibeam and rastering software were key tools that opened the door to this new class of protein structures.”
The two papers, one by Kobilka’s group and one by a group from the Scripps Research Institute led by Raymond Stevens, can be found online on Nature’s website.
The quad collimator minibeam was recognized in 2010 by R&D magazine as one of the top 100 innovations of that year.
Provided by Argonne National Laboratory