Control of gene expression: Histone occupancy in your genome
When stretched out, the genome of a single human cell can reach six feet. To package it all into a tiny nucleus, the DNA strand is tightly wrapped around a core of histone proteins in repeating units—each unit known as a nucleosome. To allow access for the gene expression machinery the nucleosomes must open up and regroup when the process is complete.
In the May 1, 2012, issue of Genes & Development, researchers at the Stowers Institute for Medical Research demonstrate how failure to restore order has lasting consequences. During the process of gene expression, a factor known as Chd1 promotes nucleosome reassembly on the DNA strand. Without it, yeast cells are unable to attach a chemical mark called ubiquitin to one of the four types of histone proteins, which in turn hampers nucleosome re-establishment throughout the entire yeast genome. In mammalian cells, this important step could be perturbed in disease states such as cancer.
"We have used the yeast Saccharomyces cerevisiae as a model system to study histone mono-ubiquitination and found that the mechanism is conserved in human cells," says Stowers Investigator Ali Shilatifard, Ph.D., who led the study. Why that's important dates back to studies initiated a decade ago on a gene called MLL, which is involved in the development of an aggressive type of leukemia in infants.
Previously, when Shilatifard's group analyzed the yeast version of MLL, they found that it enzymatically modified a histone called H3 by decorating it with a biochemical flag. In this case, that flag was not an ubiquitin, but rather a methyl group. Genes occupied by nucleosomes containing methylated H3 are generally switched on. Later, the lab confirmed that the same histone methylase activity was evolutionarily conserved from yeast to human cells, setting the stage for the molecular analysis of MLL activity in yeast.
The current study is based on a concept known as "histone crosstalk." It posits that rather than acting individually, histones within a nucleosome, particularly H3 and H2B, carry on a complex biochemical conversation among themselves based on how they are chemically modified. For example, previous studies had hinted that mono-ubiquitination at one histone (H2B) precedes methylation of another (H3), suggesting that the primary role of the former was to stimulate the latter. Thus, in this paper, Shilatifard and his team asked whether the sole function of H2B mono-ubiquitination was to regulate H3 methylation.
What Jung Shin Lee, Ph.D., a postdoctoral researcher and the study's first author, and Alexander Garrett, a bioinformatics specialist in the Shilatifard lab and co-author, found was unexpected: although yeast carrying mutant Chd1 showed a loss of H2B ubiquitination, H3 methylation was fairly unaffected. "Surprisingly, when we looked genome-wide, we observed that methylation patterns of other histones within nucleosomal complexes were fairly normal in Chd1 mutants," says Lee. "That told us that there are other functions associated with ubiquitination of H2B besides methylation of H3."
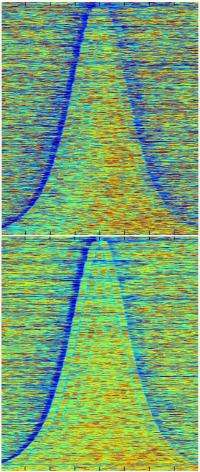
To reveal what those functions were, the investigators applied a technique that has been mastered by Penn State biochemist Frank Pugh, Ph.D. The methodology, known as nucleosomal occupancy mapping, can nail down to a single DNA base the very spot on a DNA strand where a complex of histone proteins is sitting—not in just one gene, but in all 5,700 or so yeast genes simultaneously.
Collaborating with Pugh's lab, Shilatifard's group used the technique to map loci where nucleosomes formed in the genome of normal versus Chd1-mutant yeast. Mutants showed decreased occupancy of nucleosomes compared to non-mutant cells, particularly in longer genes, suggesting that mutants have difficulty recreating the proper nucleosome array after gene expression.
"To synthesize RNA from DNA, the normal chromosome structure must be loosened to make way for RNA polymerases, enzymes that transcribe DNA," explains Garrett. "After that, the DNA must be re-condensed through the reassembly of nucleosomes." That, rather than H3 methylation, is the primary activity impaired when the protein Chd1 is nonfunctional. Finally, the investigators experimentally manipulated cultured human cells to eliminate expression of the human counterpart of Chd1—a molecular trick equivalent to analyzing Chd1-mutant yeast—and observed impaired mono-ubiquitination of human H2B, just like in yeast.
"These experiments show that the function of Chd1 that we see in single-celled yeast is conserved in humans," says Shilatifard, praising what he calls the awesome power of yeast biochemistry and genetics. "Basically, we are able to use yeast as a test tube to discover what the function of these proteins is in you and me. Given the fact that histone mono-ubiquitination is associated with processes that can lead to the development of human cancer, the molecular information obtained in yeast are indispensable for better understanding the role these factors play during human development and disease pathogenesis," says Shilatifard.
Provided by Stowers Institute for Medical Research