August 29, 2011 feature
Genes ex silico: Computer-designed virus yields phenotype expression benefits
(PhysOrg.com) -- Gene therapy is medicine’s rising star with adeno-associated virus (AAV) vectors – nonpathogenic parvoviruses – among the most promising supporting actors, due largely to their capability to integrate into transcriptionally silent genomic regions (areas that do not, via RNA polymerases, make a messenger RNA copy of DNA-stored genetic information). That being said, there’s been a downside in assembling AAV vectors into adenoviral (Ad) viral backbones, which are used extensively in genetic research and therapy: They rely on replication (Rep) proteins – in this study, the Rep 78/68 polypeptide – which limit viral amplification methodologies. Recently, however, researchers at Stony Brook University have used computational microbiology to reengineer the Rep gene through synonymous codon pair recoding, a technique in which the meaning of a codon (a set of three base nucleotides) is changed in a dynamic site- or mRNA-specific way – that is, it competes and shares expression with the standard. The experimental results demonstrate the value synonymous codon pair reengineering can have in modifying endpoint expression.
Dr. Wadie F. Bahou, MD, in Stony Brook’s Department of Medicine, works with a research group consisting of Varsha Sitaraman and Dmitri V. Gnatenko in the Department of Medicine; Patrick Hearing, Eckard Wimmer and Steffen Mueller in the Molecular Genetics and Microbiology Department; and Steve Skiena and Charles B. Ward in the Computer Sciences Department.
“Our group is composed of individuals with long-standing interests in computational biology, virology, and gene therapy using viruses,” says Bahou. Thus, my group has been most interested in modifying genomes as strategies for gene therapy, Eckard and Steffen are pioneers in recoding viruses for vaccine development, while Steve and Charles provide the critical common thread for all our computational studies.”
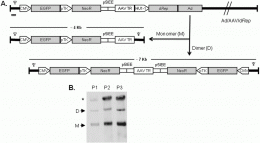
The key step in their achievement was to computationally redesign Rep 78/68 so that it was better tolerated by adenoviral vectors. “Prior research by our team had previously developed a computational algorithm for redesigning genes based on synonymous codon pair recoding,” Bahou notes. “The real leap of faith was to ask whether the algorithm could be applied to the case of viral/viral interactions. The redesign was relatively straightforward using the previously developed computational algorithm.”
The methods that the team used to demonstrate that their leap of faith was justified were equally innovative. As Bahou explains, “The real innovation here is progressive evidence that synonymous pair bias actually can be manipulated to change fundamental characteristics of biological functions. Members of our team have previously demonstrated this for poliovirus and influenza virus, and in this case, we have shown its relevance for generating a novel gene therapy adenovirus.”
Bahou also sees a number of potential avenues for further developing the group’s line of research. “Certainly the possibility of using these reengineered viral systems for vaccine development is an obvious application. For applications directly related to this most recent manuscript,” he continues, “it provides a unique opportunity for safe methods for long-term gene delivery. On a broader application, one could envision entire cell systems with recoded genes as a means of altering cellular function in general. This is a forward-thinking statement, but certainly predicated on the status of genome synthesis coupled with progressive information on genetic pathways that regulate cell functions.”
Those applications, he adds, will most likely feature the ability to rapidly characterize and synthesize attenuated genomes as new vaccines. In addition, “we are currently testing our ability to safely use this technology for gene replacement strategies which have been limited by concerns for insertional mutagenesis” – mutations caused by inserting genes into a genome. “Such an issue is especially relevant for benign diseases such as hemophilia where cellular transformation using integrating lentiviruses should be considered an unacceptable risk for long-term gene replacement strategies. Therefore,” Bahou points out, “our collective group is currently trying to apply this research as a safe means of long-term gene replacement.
At the same time, Bahou stresses that “It’s important to note that genome reengineering using synonymous codon pair restructuring is a relatively new concept, and we fully anticipate that the applications will broaden over time. In theory,” he concludes, “we should be able to modify genomes for both attenuated expression and enhanced expression, depending on the situational requirements. In that respect, the technology is relevant for many other biological systems, especially when developing computational approaches to modifying cellular systems at many levels.”
More information: Computationally designed adeno-associated virus (AAV) Rep 78 is efficiently maintained within an adenovirus vector, Published online before print August 15, 2011, doi:10.1073/pnas.1102883108 , PNAS August 23, 2011 vol. 108 no. 34, 14294-14299
Copyright 2011 PhysOrg.com.
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.