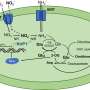
A recent study into the interactions between aminoglycoside antibiotics and their target site in bacteria used computer simulations to elucidate this mechanism and thereby suggest drug modifications.
In the article, which will be published on July 21st in the open-access journal PLoS Computational Biology, researchers from University of Warsaw, Poland, and University of California San Diego, USA, describe their study of the physical basis of one bacterial resistance mechanism - mutations of the antibiotic target site, namely RNA of the bacterial ribosome. They performed simulations and observed changes in the interaction between the antibiotic and the target site when different mutations were introduced.
In hospitals throughout the world, aminoglycosidic antibiotics are used to combat even the most severe bacterial infections, being very successful especially against tuberculosis and plague. However, the continuous emergence of resistant bacteria has created an urgent need to improve these antibiotics. Previous experiments on bacteria have shown that specific point mutations in the bacterial ribosomal RNA confer high resistance against aminoglycosides. However, the physico-chemical mechanism underlying this effect has not been known. Using computer simulations the researchers explained how various mutations in this specific RNA fragment influence its dynamics and lead to resistance.
Bacteria have developed other ways of gaining resistance, not just through mutations, and further studies are underway. The authors are now investigating the resistance mechanism by which bacterial enzymes actively modify and neutralize aminoglycosidic antibiotics. These molecular modeling studies together with experiments could help to design even better aminoglycoside derivatives in the future.
More information: Romanowska J, McCammon JA, Trylska J (2011) Understanding the Origins of Bacterial Resistance to Aminoglycosides through Molecular Dynamics Mutational Study of the Ribosomal A-Site. PLoS Comput Biol 7(7): e1002099. doi:10.1371/journal.pcbi.1002099
Provided by Public Library of Science