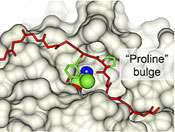
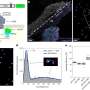
Molecular structure of ISP protease
(PhysOrg.com) -- A Cardiff-led team has found a unique type of protein inside bacterial cells which could shed new light on organisms such as the disease-causing C. difficile.
A class of enzyme, known as the subtilisin proteases, are found in every form of life from viruses and bacteria to humans. Normally, these proteases only act outside the cell – never inside it.
A team from the School of Biosciences, working with colleagues in York, have identified a class of subtilisin proteases that work inside the cell. They have characterised the molecular three-dimensional structures of the proteases, showing up unique features compared to proteases operating outside the cell only.
Dr Dafydd Jones of the School of Biosciences, who led the study, said: "The active components of cells are comprised of protein – which proteases normally attack. So, to have an active protease inside the cell is potentially very disruptive. However, a variety of different bacteria have this protease inside the cell and are able to regulate its activity."
Molecular analysis by the team found a section of the protease bent back over its active part, limiting its effect within the cell. The next challenge is to work out exactly what function the protease carries out within the bacteria.
Dr Jones said: "This protease is only found in certain bacteria types, including the well-known C. difficile and B. anthracis. We know that subtilisins outside the cell play a key role in many different functions, such as digestion of food and the regulation of hormones. If we could work out the precise function of this new class of protein, it could tell us a great deal about the life cycle of these various bacteria."
The research team also included Michael Gamble and Georg Künze of the School of Biosciences. The findings have just been published in the leading journals Structure and PNAS.
Provided by Cardiff University