New crystallization method to ease study of protein structures
Researchers at the Midwest Center for Structural Genomics (MCSG), the Structural Genomics Consortium (SGC) and the Structural Biology Center (SBC) at the U.S. Department of Energy's Argonne National Laboratory have developed a new technique for crystallizing proteins that will ease experimentation into protein structures.
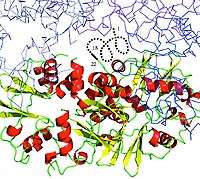
In order to study protein structures, biologists must turn what is essentially a soup of purified protein into crystals that have a consistent and ordered structure. Each protein consists of a chain of amino acid subunits that twists into helices, ribbons and loops. Some proteins have less tidy molecular structures than others; in these, disordered amino acid chains dangle off the protein like split ends.
Crystallizing proteins that contain many of these flexible regions takes much more work and patience than working with more organized ones, said Argonne senior biologist Andrzej Joachimiak, who led the Argonne research effort. "We've tried to find a way to remove the disordered parts using computer modeling, but that's been a challenging process," he said. "This new experimental method is fast, inexpensive and can be applied to many different targets, from bacterial pathogens to human proteins."
In order to try to boost the efficiency of the crystallization process, Joachimiak and his colleagues at the MCSG and SGC inserted a protease—a certain type of enzyme that breaks down the bonds that connect a protein's amino acids.
Once added, the protease preferentially bound to the proteins at the disordered regions, snipping off the loose ends like a molecular barber. The researchers successfully crystallized and examined nine of these newly shorn proteins that previously had resisted attempts to study them using X-ray crystallography.
"This simple technique offers an opportunity to uncover and characterize the structures of dozens of proteins that up until now we had to study using much more laborious and expensive approaches," Joachimiak said.
This process, known as "limited in situ proteolysis," represents one of several potential "salvage pathways" that biophysicists could use to create more usable protein crystals and reduce waste, Joachimiak said. Currently, scientists' efforts to manufacture and then study a workable crystal on Argonne's Advanced Photon Source yield structural data only about 15 percent of the time. By using proteases to digest part of the protein sample, the Argonne scientists achieved a six percent boost in efficiency.
Joachimiak cautioned that scientists do not have a way to successfully crystallize every protein, even with the use of proteolysis. "There will still be some that are resistant," he admitted, "but we are making enormous strides in our understanding of how exactly these essential substances work."
A research paper, "In situ proteolysis for protein crystallization and structure determination," that detailed the study appeared in the December 4 issue of Nature Methods. The study's X-ray data were collected at the SBC beamlines at the Advanced Photon Source. The MCSG and SGC represent a collaboration of Argonne scientists as well as scientists from Canada and Europe.
Source: Argonne National Laboratory