Blending bacterial genomes for megacloning
Scientists in Tokyo report developing a "megacloning" method of transferring entire genomes from one bacterial species into another.
DNA bacterial cloning is routinely used to isolate genes and understand gene function, but most bacteria cannot handle a large number of genes at one time.
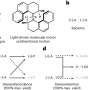
In an attempt to increase the genomic size for DNA cloning, Mitsuhiro Itaya and colleagues from the Institute of Life Sciences made use of a DNA cloning vector derived from the 4.2-Mb sized genome of the Bacillus subtilis bacteria, and for the target DNA used the 3.5-Mb genome of the non-pathogenic, photosynthetic bacterium Synechocystis.
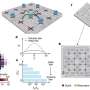
The researchers divided the Synechocystis genome into four regions of approximately equal size and guided that target DNA into the cloning vector by genetic recombination at "landing pad sequences."
Through progressive assembly and editing of DNA regions covering the entire Synechocystis genome sequence, the resultant bacteria contained a composite genome size of 7.7 Mb.
The scientists said their megacloning method extends size limits of stable DNA cloning and might lead to construction of beneficial microbes for bioindustrial use.
Their research appears in the online early edition of the Proceedings of the National Academy of Sciences.
Copyright 2005 by United Press International