Bacteria control how infectious they become, study finds
The results of a new study suggest that bacteria that cause diseases like bubonic plague and serious gastric illness can turn the genes that make them infectious on or off.
Knowing how disease-causing bacteria, like Yersinia pestis and E. coli, do this may one day help scientists create drugs that control the expression of these genes, thereby making the bacteria harmless, said Vladimir Svetlov, a study co-author and a research associate in microbiology at Ohio State University. The findings appear in the April 13 issue of the journal Molecular Cell.
Gene expression – the process of turning on, or activating, genes – is controlled by proteins called transcription factors. Every type of bacteria known to humankind contains the transcription factor NusG, which controls nearly all of a bacterium's gene expression. Without it, a microorganism will die.
“We think that NusG regulates nearly every gene in every form of bacteria,” said Irina Artsimovitch, the study's lead author and an associate professor of microbiology at Ohio State . “Say a bacterium has 3,000 genes – NusG would regulate 2,900 of them.”
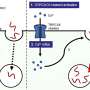
But somewhere along the evolutionary path, NusG was copied and physically changed. The result was a specialized transcription factor called RfaH. Unlike NusG, RfaH controls only a small portion of gene expression. But it happens to turn on those genes that give bacteria like E. coli and Y. pestis their ability to infect.
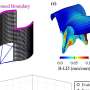
The researchers say that this study likely marks the first successful attempt by a laboratory to determine the structure of RfaH.
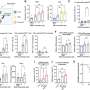
They used special X-ray techniques to study and describe RfaH proteins that they had extracted from E. coli. They found that while about two-thirds of RfaH's structure closely resembles the structure of NusG, the remaining one-third looked dramatically different. It's this latter third that appears to be the portion of the protein responsible for controlling the genes that make E. coli infectious.
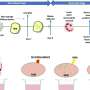
“In contrast to NusG, which is always active, RfaH is usually inactive, because the part of the protein that is needed to activate gene expression is typically masked,” Svetlov said.
It's only when RfaH finds the appropriate target sequence on a bacterium's DNA that this small portion of the protein is unmasked and can then turn on a select group of genes. These genes let disease-causing bacteria infect their host while at the same time protecting the bacteria from the host's immune defenses.
“E. coli seems to prevent RfaH from acting unless the microorganism absolutely needs it,” Artsimovitch said. That's because bacteria like E. coli are caught in a delicate balancing act. With too little RfaH, bacteria grow too slowly. But too much RfaH, and they will die.
While RfaH's control over gene expression is limited, it seems that its structure lets it control key sequences of the genome during transcription, the process of transferring genetic information inside a cell and one of the first steps of gene expression.
“Making RfaH work only at specific sites is, in a sense, a genius way to prevent it from interfering with NusG,” Artsimovitch said. “It seems that the only genes that RfaH can't regulate are those controlled by NusG.”
Bacteria can survive without RfaH, but not without NusG. Yet without RfaH, bacteria lose the ability to infect. In previous laboratory experiments, the researchers found that pathogens lacking RfaH grow at much slower rates.
“Cells usually don't die when RfaH use changes,” Svetlov said. “Rather, bacteria seem to manipulate the protein, to play around with it. Too much RfaH will kill a cell, while too little would prevent it from infecting any living being.
“We think that RfaH is responsible for more than making a microbe infectious,” he continued. “Actually seeing what happens at the molecular level will help us figure out what else this protein regulates.”
Svetlov and Artsimovitch conducted the study with Georgy Belogurov, a postdoctoral research associate in microbiology at Ohio State, and with researchers from the University of Alabama at Birmingham, the Howard Hughes Medical Institute and the University of Texas Southwestern Medical Center in Dallas.
Source: Ohio State University