Zooming in on a Molecular Relay
In the cellular milieu, a protein is only as good as the tags it wears. If a protein is flashing a specific type of tag, it is marked for destruction, whereas another tag might signal a green-light for cell division.
Howard Hughes Medical Institute researcher Brenda Schulman has been fascinated with how these tags are generated and how they help get proteins ready for their cellular jobs. One of her broad goals is to learn how cells respond quickly to the changing demands and cues of their environments.
Writing in the current issue of Nature, Schulman and her colleagues at St. Jude Children's Research Hospital have taken a large step toward that goal by learning how a set of enzymes contorts itself during the molecular relay race involved in tagging proteins. These tags, known as ubiquitin-like proteins, are attached to specific proteins in the cell. The set of enzymes, which work together to find, prepare, escort, and attach these tags to their assigned target molecules, can control the fate of individual proteins and, ultimately, a cell.
Attaching ubiquitin or a ubiquitin-like tag to a protein can trigger a protein's destruction or launch a specific cellular activity, such as one of the steps of cell division. To keep cells healthy, it is essential that the right molecules are tagged at the right time - and it's the job of a large family of enzymes known as E1, E2, and E3 to see that they do. Deregulation of the system has been implicated in cancers, neurodegenerative disorders, and some viral infections. “The pathways by which ubiquitin-like proteins are attached to target proteins are essential for a broad array of cell processes,” explained Schulman. “These processes include cell division, embryonic development and the immune response. Malfunction of these processes can lead to numerous diseases,” she said.
Schulman and her colleagues reported new details of the E1-E2-E3 enzyme “cascades” that attach ubiquitin and more than a dozen ubiquitin-like proteins to their targets in an article in an advance online publication on January 14, 2006, in the journal Nature.
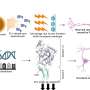
E1, E2, and E3 enzymes catalyze a complex series of reactions, with each enzyme playing a distinct role. After identifying the correct ubiquitin-like protein, E1 enzymes activate it by attaching it to the enzyme via a sulfur-containing structure called a thioester. Next, the thioester link is transferred to a second enzyme called E2—handing off the ubiquitin (or other tag) much as one relay runner hands off a baton to another. Finally, the E2 enzyme usually associates with an enzyme called E3, which stitches the ubiquitin to its target protein.
Schulman's lab has been working to understand the molecular basis of this highly coordinated, many-step process. Their structural biology studies have helped reveal how E1 enzymes recognize and activate specific ubiquitin-like proteins. In the new study reported in Nature, they wanted to explore in more detail how E1 passes off the ubiquitin-like protein to the next enzyme in the pathway.
The number of E1-E2-E3 enzyme cascades that attach ubiquitin to its thousands of targets within the cell is enormous. To make their task more manageable, Schulman and her colleagues studied the more straightforward E1-E2-E3 cascade that attaches a ubiquitin-like protein called NEDD8, whose function is very similar to ubiquitin. Thus, the mechanism of its cascade would be essentially the same, said Schulman. Since the steps in the reaction cascade are so transient, Schulman and her colleagues studied the transfer process by genetically altering the E2 enzyme such that it would trap the process in mid-reaction—like freezing two relay runners in mid-handoff.
The researchers used x-ray crystallography to study the conformation of the trapped enzymes. In x-ray crystallography, researchers bombard protein crystals with x-ray beams. As the x-rays pass through and bounce off of atoms in the crystal, they leave a diffraction pattern, which can then be analyzed to determine the three-dimensional shape of the protein.
Analyzing this structure, the researchers discovered that after E1 binds to NEDD8, E1 alters its conformation to allow E2 to face in the right direction to accept the NEDD8 handoff. “The real key here is that the ubiquitin-like protein getting transferred is the basis of the switch,” said Schulman. This “thioester switch” triggers the transfer process that includes the shifting of the NEDD8 to E2 and the release of E2 from E1, she said.
“This work helps solve a fundamental puzzle about the cascade process,” said Schulman. “Previous models in the field indicated that the enzymes were oriented like a baton handoff by two professional relay runners, in which the second was facing away from the first. But given what we knew previously about the transfer cascade, that orientation made no sense; the enzymes were not in the proper relationship to one another. Our new data reconciles previous models with the physical requirements of the handoff. We have shown that the handoff occurs as if between two inexperienced relay runners, in which the second turns around to face the first for the handoff. But after the handoff, the enzymes would reorient so that the second faces away from the first, to separate and gain distance, just as in a real relay race.” According to Schulman, the mechanism of transfer of bonds between such enzymes that she and her colleagues discovered may well be a fundamental one for such processes.
The next objectives of her research, she said, will be to understand the subsequent reactions that involve the stitching of a ubiquitin-like protein to its target protein by E3, as well as how the ubiquitin-like protein is recognized by cellular machinery that directs proteins to the garbage heap or to their functional destination.
Schulman collaborated on the studies with researchers from Harvard Medical School and Lawrence Berkeley National Laboratory.
Source: Howard Hughes Medical Institute