Chemists create 'nanorobotic' arm to operate within DNA sequence
New York University chemistry professor Nadrian C. Seeman and his graduate student Baoquan Ding have developed a DNA cassette through which a nanomechanical device can be inserted and function within a DNA array, allowing for the motion of a nanorobotic arm. The results, reported in the latest issue of the journal Science, mark the first time scientists have been able to employ a functional nanotechnology device within a DNA array.
“It is crucial for nanorobotics to be able to insert controllable devices into a particular site within an array, thereby leading to a diversity of structural states,” explained Seeman. “Here we have demonstrated that a single device has been inserted and converted at a specific site.”
He added that the results pave the way for creating nanoscale “assembly lines” in which more complex maneuvers could be executed.
The results are based upon a device Seeman and his NYU colleagues had previously developed. That component has enabled the translation of DNA sequences, thereby potentially serving as a factory for assembling the building blocks of new materials. The invention has the potential to develop new synthetic fibers, advance the encryption of information, and improve DNA-based computation.
The device, developed with NYU Chemistry graduate student Shiping Liao, emulates the process by which RNA replicas of DNA sequences are translated to create protein sequences. However, the signals that control the nanomechanical tool are DNA rather than RNA. The dimensions of the machine are approximately 110 x 30 x 2 nm.
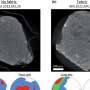
In this study, Seeman and Ding developed a framework that contains a binding site--a cassette—that allows insertion of the device into a specific site of a DNA array. Changing the cassette’s control sequences or insertion sequences would allow the researchers to manipulate the array or insert it at different locations. The researchers added a long arm to the framework so that they could observe the structure undergoing a half-rotation. They visualized their results by atomic force microscopy (AFM), which permits features that are a few billionths of a meter to be visualized.
Source: New York University