Biobarcode Nanoparticles Enable Multiplexed DNA Detection
Given that cancer is a disease that results from gene mutations, the development of high-throughput schemes for detecting specific DNA sequences would have a dramatic effect on cancer research, cancer detection, and the monitoring of therapeutic efficacy.
And while DNA microarrays have proven themselves as useful tools for such high-throughput mutation detection assays, a variety of technical limitations suggests that researchers need to develop faster, simpler, and less expensive methods for identifying cancer-related gene mutations.
Gold nanoparticles containing DNA "barcodes" may provide that next generation technology. Writing in the journal Angewandte Chemie International Edition, Chad Mirkin, Ph.D., and his colleagues at Northwestern University show how biobarcoded nanoparticles, which up until now have proven useful only for detecting mutations one at a time, can be used in a multiplexed system to detect and distinguish among four different DNA sequences simultaneously. In this case, the investigators used DNA from four different viruses as their model system, but they note that their results apply to any set of unique DNA sequences.
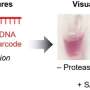
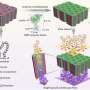
The investigators began their experiments by constructing biobarcodes, nucleic acid sequences of 30 to 33 bases. Part of each biobarcode recognizes a specific target DNA sequence, while the remainder of each biobarcode is common among all barcodes and is necessary for detection and readout functions in the assay. Each biobarcode is linked to a 30-nanometer-diameter gold nanoparticle. The researchers also constructed magnetic microparticles containing a short piece of DNA that binds to a separate unique region of the target DNA.
The researchers were able to use these components to successfully identify each of the four viral DNA samples in various mixtures of viral DNA. The test results showed that enzymatic target amplification, using polymerase chain reaction, for example, was not needed to correctly identify even low levels of the target DNA. The investigators note that because it is possible to create billions of different biobarcode sequences, it should be possible to extend this assay system to detect and identify myriad gene mutations, such as those that should be identified by The Cancer Genome Atlas (TCGA) project.
This work is detailed in a paper titled, "Multiplexed DNA detection with biobarcoded nanoparticle probes." Mirkin is the principal investigator of Northwestern University's Center of Cancer Nanotechnology Excellence. An abstract of this paper is available through PubMed.
Source: National Cancer Institute