Decoding chronic lymphocytic leukemia
A paper published online on June 13 in the Journal of Experimental Medicine identifies new gene mutations in patients with chronic lymphocytic leukemia (CLL) -- a disease often associated with lack of response to chemotherapy and poor overall survival.
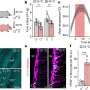
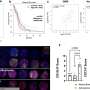
CLL is the most common leukemia in the Western world, but the disease varies greatly from patient to patient with regard to prognosis, survival, and disease course. In attempt to understand the genetic basis for this heterogeneity, a group led by Riccardo Dalla-Favera at Columbia University and Gianluca Gaidano at Amedeo Avogadro University of Eastern Piedmont, Novara, Italy surveyed the landscape of mutations in the genes of CLL patients. They found several mutations not previously linked with CLL, but most patients had relatively few genetic mutations compared to some other types of cancer.
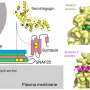
The team then turned their attention to NOTCH1, a gene that controls cell division and survival and is often activated by mutation in other types of leukemia. CLL patients harboring NOTCH1 mutations at the time of diagnosis had a significantly poorer prognosis and shorter survival than patients without NOTCH1 mutations. And NOTCH1 mutations were much more common in samples from patients whose disease progressed to more high-risk forms or who failed chemotherapy treatment.
These findings point to the NOTCH1 pathway as potentially useful for diagnostic and therapeutic purposes in human CLL.
More information: Fabbri, G., et al. 2011. J. Exp. Med. doi:10.1084/jem.20110921