Researchers advance understanding of structural change in cancer cells
(PhysOrg.com) -- A method developed by a University of Maine mathematician to get a much more detailed look at cellular morphology has the potential to aid in early cancer detection.
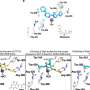
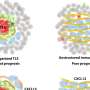
Andre Khalil, a UMaine assistant professor of mathematics and cooperating assistant professor of physics, has developed a computational algorithmic method for characterizing three-dimensional images of cellular nuclei. Using new analysis and measurement algorithms, the procedure can quantify for the first time in unprecedented detail the size, shape, position and proximity of chromosomes in cell nuclei, a breakthrough in creating an extensive new baseline database of cellular structure.
Khalil, who works in collaboration with cancer researcher Kevin Mills of The Jackson Laboratory in Bar Harbor, Maine, and colleagues in Europe, is creating one of the world’s largest collection of chromosomal images, called chromosome territories, that will feed a database for comparing normal and abnormal cells. The comparisons can reveal a chromosome’s changing shape and architecture, which could indicate the onset of one of the five stages of tumor development.
Currently, the database contains up to 700 cellular images from The Jackson Laboratory and more than 20,000 are being donated by cooperating researchers in England, France, Germany, Canada and the United States.
Learning how and when chromosome and nuclear structure in cells begin to change with the onset of cancer could help scientists determine which biophysical and genomic mechanisms cause the changes. The question is whether these mechanisms can be targeted, controlled, treated or dampened, Khalil says.
“Not only do we want to quantify changes between normal and cancerous cells, but how early in the stage can we quantify it,” says Khalil, whose research support includes nearly $80,000 from the Maine Cancer Foundation for 2010-2011. “The structural changes we are looking to rigorously quantify are the hallmark of many types of cancer.”
Khalil, a collaborator with the Chromatin and Genome Research Group at the Ecole Normale Superieure de Lyon in France, also conducts research that compares breast lesions and tumors in an effort to detect cancerous cells. His cellular and chromosomal analysis involves lymphoid cancer cells. The cell nucleus image database now under development for early detection in blood cancers such as lymphoma can also be applied to detect other types of cancer.
In his image analysis and computational modeling lab at UMaine, Khalil has pioneered several novel applications of the WTMM (Wavelet Transform Modulus Maxima) method, an image and signal processing technology — a mathematical microscope, if you will — in astrophysics and biomedicine. Some of those applications include analysis of soft-tissue growth and artificial bone implants, breast cancer tumor analysis, and algorithmically refining signals from sleeping infants to detect interrupted sleep.
A previous discovery Khalil made with fluorescent microscopy — which creates two-dimensional or three-dimensional images of a cell nucleus — established for the first time that, contrary to common thinking, chromosomes are not spherical, but elongated and surfboard-shaped.
The work is particularly relevant for Maine, where the cancer rate has been the highest in the nation at 510 cases per 100,000 residents in 2005, according to the U.S. Cancer Statistics Report. The national rate was 460 cases per 100,000 people, the Report says.
The development of Khalil’s computational tools, Khalil adds, could be used for cytological analysis for colorectal, esophageal, bladder, and lung cancer, of which Maine has an unusually high incidence, he says.