Kinked nanopores slow DNA passage for easier sequencing

(PhysOrg.com) -- In an innovation critical to improved DNA sequencing, a markedly slower transmission of DNA through nanopores has been achieved by a team led by Sandia National Laboratories researchers.
Solid-state nanopores sculpted from silicon dioxide are generally straight, tiny tunnels more than a thousand times smaller than the diameter of a human hair. They are used as sensors to detect and characterize DNA, RNA and proteins. But these materials shoot through such holes so rapidly that sequencing the DNA passing through them, for example, is a problem.
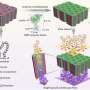
In a paper published this week online (July 23) in Nature Materials (hardcopy slated for August, Vol.9, pp. 667-675), a team led by Sandia National Laboratories researchers reports using self-assembly techniques to fabricate equally tiny but kinked nanopores. Combined with atomic-layer deposition to modify the chemical characteristics of the nanopores, the innovations achieve a fivefold slowdown in the voltage-driven translocation speeds critically needed in DNA sequencing. (Translocation involves DNA entering and passing completely through the pores, which are only slightly wider than the DNA itself.)
"By control of pore size, length, shape and composition," says lead researcher Jeff Brinker, "we capture the main functional behaviors of protein pores in our solid-state nanopore system." The importance of a fivefold slowdown in this kind of work, Brinker says, is large.
Also of note is the technique's capability to separate single- and double-stranded DNA in an array format. "There are promising DNA sequencing technologies that require this," says Brinker.
The idea of using synthetic solid-state nanopores as single-molecule sensors for detection and characterization of DNA and its sister materials is currently under intensive investigation by researchers around the world. The thrust was inspired by the exquisite selectivity and flux demonstrated by natural biological channels. Researchers hope to emulate these behaviors by creating more robust synthetic materials more readily integrated into practical devices.
Current scientific procedures align the formation of nominally cylindrical or conical pores at right angles to a membrane surface. These are less capable of significantly slowing the passage of DNA than the kinked nanopores.
"We had a pretty simple idea," Brinker says. "We use the self-assembly approaches we pioneered to make ultrathin membranes with ordered arrays of about 3-nanometer diameter pores. We then further tune the pore size via an atomic-layer deposition process we invented. This allows us to control the pore diameter and surface chemistry at the subnanometer scale. Compared to other solid state nanopores developed to date, our system combines finer control of pore size with the development of a kinked pore pathway. In combination, these allow slowing down the DNA velocity."
The work is supported by the Air Force Office of Scientific Research, the Department of Energy's Basic Energy Sciences and Sandia's Laboratory Directed Research and Development office.
More information: The paper is slated to be published hardcopy in August 2010 (Vol.9, pp. 667-675) of Nature Materials.
Provided by Sandia National Laboratories