Gently unfolding proteins to watch them refold
(PhysOrg.com) -- How does a protein chain fold into the same 3-D shape each time and not something disfunctional or dangerous? A new study shows that the first fold is critical. The finding by Susan Marqusee and Carlos Bustamante could help to understand the role of misfolded proteins in Alzheimer's Disease.
Alzheimer’s disease belongs to a family of conditions classified as amyloidoses, in which misfolded proteins clump into plaques or fibrils instead of taking their normal, soluble form. Why this happens and how it might be prevented has been the subject of intense research for decades. Equally of interest is how most proteins have evolved to avoid misfolding.
QB3 researchers Susan Marqusee, a UC Berkeley professor of biochemistry and molecular biology, and Carlos Bustamante, a UC Berkeley professor of physics, show in new research that, in T4 lysozyme—an enzyme related to one responsible for a form of amyloidosis—a subtle detail of topology ensures that the protein either folds all at once or not at all, avoiding dangerous intermediates.
The research was published in the journal Nature. First authorship was shared by Elizabeth Shank, now a postdoctoral fellow at Harvard; Ciro Cecconi, now a researcher at the University of Modena and Reggio Emilia in Italy; and Jesse Dill, a graduate student at UC Berkeley.
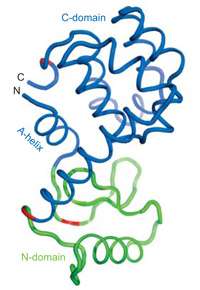
T4L, like all proteins, is a string of amino acid residues. At the “C” end is a carboxylic acid group; at the “N” end, an amino group. When folded, T4L consists of two globular domains. Starting from the N end, the first 12 residues coil into what the authors call the “A-helix.” The next 48 residues comprise the N-domain; the last 104 residues belong to the C-domain. But the initial A-helix folds into the C-domain.
“This ‘re-entrant’ structure is found in many enzyme families where the active site is in the cleft between domains,” Marqusee says.
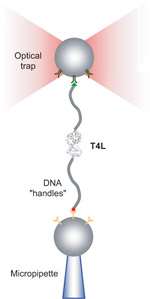
The researchers used a technique called “laser tweezers” to gently pull single T4L enzymes apart and allow them to refold over and over again. Single molecules are subject to random thermal fluctuations, which induce variations in the curves of force versus extension. From these variations, the scientists used a sophisticated mathematical technique to extract information about the “free energy” difference between folded and unfolded T4L.
What they found was that pulling apart just the N-domain required forces and energies similar to those required to unfold the entire enzyme, implying that the two domains folded “cooperatively”—the C-domain could not fold independently of the N-domain.
T4L’s structure has an unusual quirk: folded, its N end and C end sit next to each other. This enabled the researchers to genetically engineer a version in which the A-helix was cut from the N end and joined to the C end, so that there was only one point at which the two domains made contact. Experiments showed that the domains of this engineered T4L folded independently.
The results are evidence that T4L’s A-helix is a sort of Velcro reinforcing strip holding the protein together. “We speculate that the re-entrant structure in many proteins has been selected over evolutionary time to ensure that different domains fold cooperatively,” Bustamante says. "This prevents misfolding,” known to be a major factor in amyloidosis.
The new results could be useful to scientists designing enzymes from scratch, Marqusee says.
“Our collaboration is a good example of the kind of work that can emerge from the kind of multidisciplinary environment fostered by QB3,” Bustamante says. “By combining molecular biology and physics, we have been able to discover communication between the domains of the proteins that would have been difficult to extract in any other way.”
“This is beautiful work,” says Nancy Forde, an assistant professor of physics at Simon Fraser University in Vancouver, who was not involved in the research. “The Marqusee and Bustamante groups have made seminal contributions in two distinct fields: further demonstrating how optical tweezers can provide unique insight into protein structure and stability, and illustrating the critical influence of re-entrant structure on the folding pathways of an enzyme.”
More information: Nature paper: www.nature.com/nature/journal/ … ull/nature09021.html
Provided by University of California - Berkeley