Effects of lifestyle and exposures are mirrored in blood gene expression
A study by Norwegian and French researchers hopes to provide new understanding of how blood cells adjust gene expression in response to various clinical, biochemical and pathological conditions. The Norwegian Woman and Cancer (NOWAC) postgenome study, published March 12 in the open-access journal PLoS Genetics, highlights numerous blood gene sets affected by one's physical condition, lifestyle factors and exposure variables.
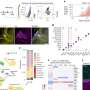
The authors used 286 blood samples from postmenopausal Norwegian women, collected from the NOWAC postgenome biobank and processed via both a standardized blood collection procedure and an experimentally-validated microarray platform. Using these samples, the researchers investigated blood gene expression changes due to technical variability (e.g., how samples were collected, transported, and stored), normal inter-individuality (e.g., body mass index), and exposure variables (e.g., smoking, hormone therapy, and medication use), at levels relevant to real life situations. They established that these effects are mirrored in the blood.
Although blood gene expression profiling promises molecular-level insight into disease mechanisms, there remains a lack of baseline data describing the nature and extent of variability in blood gene expression in the general population. Characterizations of this variation and the underlying factors that most influence gene expression among healthy individuals play an important role in the feasibility, design and analysis of future blood-based studies investigating biomarkers for exposure, disease progression, diagnosis or prognosis.
The authors conclude that their findings establish the feasibility of blood gene expression profiling to detect exposure-specific differences in the general population, and that failure to consider this type of technical or biological variation can result in the misidentification of genes when investigating predictive, diagnostic or prognostic signatures in blood.
More information: Dumeaux V, Olsen KS, Nuel G, Paulssen RH, Børresen-Dale A-L, et al. (2010) Deciphering Normal Blood Gene Expression Variation—The NOWAC Postgenome Study. PLoS Genet 6(3): e1000873. doi:10.1371/journal.pgen.1000873