Scientists visualize how bacteria talk to one another

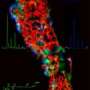
Using imaging mass spectrometry, researchers at the University of California, San Diego have developed tools that will enable scientists to visualize how different cell populations of cells communicate. Their study shows how bacteria talk to one another - an understanding that may lead to new therapeutic discoveries for diseases ranging from cancer to diabetes and allergies.
In the paper published in the November 8 issue of Nature Chemical Biology, Pieter C. Dorrestein, PhD, assistant professor at UC San Diego's Skaggs School of Pharmacy and Pharmaceutical Sciences, and colleagues describe an approach they developed to describe how bacteria interface with other bacteria in a laboratory setting. Dorrestein and post-doctoral students Yu-Liang Yang and Yuquan Xu, along with Paul Straight from Texas A&M University, utilized technology called natural product MALDI-TOF (Matrix Assisted Laser Desorption Ionization-Time of Flight) imaging mass spectrometry to uniquely translate the language of bacteria.
Microbial interactions, such as signaling, have generally been considered by scientists in terms of an individual, predominant chemical activity. However, a single bacterial species is capable of producing many bioactive compounds that can alter neighboring organisms. The approach developed by the UCSD research team enabled them to observe the effects of multiple microbial signals in an interspecies interaction, revealing that chemical "conversations" between bacteria involve many signals that function simultaneously.
"Scientists tend to study the metabolic exchange of bacteria, for example penicillin, one molecule at a time," said Dorrestein. "Actually, such exchanges by microbes are much more complex, involving 10, 20 or even 50 molecules at one time. Now scientists can capture that complexity."
The researchers anticipate that this tool will enable development of a bacterial dictionary that translates the output signals. "Our ability to translate the metabolic output of microbes is becoming more important, as they outnumber other cells in our body by a 10 to one margin," Dorrestein explain. "We want to begin to understand how those bacteria interact with our cells. This is a powerful tool that may ultimately aid in understanding these interactions."
In order to communicate, bacteria secrete molecules that tell other microbes, in effect, "I am irritated, stop growing," "I need more nutrients" or "come closer, I can supply you with nutrients." Other molecules are secreted that may turn off the body's defense mechanisms. The team is currently mapping hundreds of such bacterial interactions. Their hope is that this approach will also enable them to translate these bacterial-mediated mechanisms in the future.
Understanding the means by which microorganism cells talk to one another will facilitate therapeutic discovery, according to Dorrestein. For instance, knowing how microbes interact with human immune cells could lead to discovery of novel immune system modulators, and how these molecules control bacterial growth may lead to new anti-invectives. Both are active areas of investigation in his laboratory.
Source: University of California - San Diego (news : web)