Parasites in the genome -- A molecular parasite could play an important role in human evolution
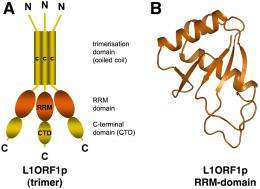
Researchers at the Max Planck Institute for Developmental Biology in Tübingen, Germany, determined the structure of a protein (L1ORF1p), which is encoded by a parasitic genetic element and which is responsible for its mobility.
The so-called LINE-1 retrotransposon is a mobile genetic element that can multiply and insert itself into chromosomal DNA at many different locations. This disturbs the genetic code at the site of integration, which can have serious consequences for the organism. On the other hand, this leads to genetic variation, an absolute prerequisite for the evolution of species. The structure of the L1ORF1p protein now allows a much more precise investigation of the mechanism of LINE-1 mobilization. This provides new insight into the relation between retrotransposons and retroviruses and probably also into certain evolutionary processes in humans and animals.
Moreover, the researchers assume that the mechanism of LINE-1 retrotransposition can be exploited one day to precisely insert genetic information into specific locations. This would be an alternative to contemporary, less location-specific methods that are based on a retroviral mechanism. (PNAS, January 20th, 2009)
The LINE-1 retrotransposon is a mobile gene that has multiplied massively in the history of the human genome. Presently, approximately 17 per cent of our DNA consist of LINE-1 sequences. This is an enormous proportion if one considers that the roughly 30.000 human proteins are encoded by less that 5 per cen of the DNA. The LINE-1 retrotransposon not only propagates itself, but also is responsible for the genomic integration of approximately one million Alu-sequences (another parasitic gene). Alu-sequences are only present in higher primates and occupy another 10 per cent of our genome. The insertion of LINE-1 and Alu sequences is a continuous process and roughly every twentieth newborn is estimated to contain at least one new insertion of such an element. Consequently, there rarely is a human gene that has not been affected in the past by the integration of a LINE-1 or Alu element.
“It is difficult to believe that the massive integration of LINE-1 and Alu sequences remained without consequences on human evolution. Thus it is surprising how little we know so far about the mechanism of retrotransposition and about the proteins and nucleic acids involved in this process“, says Oliver Weichenrieder, leading scientists at the Max Planck Institute for Developmental Biology. The researchers therefore try to gain new insights via the biochemical characterisation of the involved molecules and via the determination of their molecular structures. This provides the basis for a detailed functional analysis and reveals similarities to already known proteins, especially similarities that are not obvious from a simple comparison of the respective amino acid sequences.
In the presently published work Elena Khazina und Oliver Weichenrieder characterize one of two proteins that are encoded by the human LINE-1 retrotransposon. This so-called L1ORF1p protein binds to LINE-1 RNA, which was transcribed from a LINE-1 element in the genomic DNA. Subsequently, L1ORF1p likely supports the following reverse transcription of LINE-1 RNA into DNA. This process happens directly at the genomic integration site of the new LINE-1 element. The researchers show that the L1ORF1p protein consists of three parts. The first part causes a self-association such that always three molecules come together to form a trimer. The other two parts are necessary for binding LINE-1 RNA. “Especially surprising was the identification of a so-called RRM domain in the middle part of the protein, since this part was believed so far to be rather unstructured”, says Elena Khazina. “Our crystal structure clearly proves the existence of this domain. Meanwhile we identified RRM-domains also in other retrotransposons, in a variety of animal and plant species“, adds the structural biologist.
RRM-domains (RNA Recognition Motif) occur frequently in the cell, particularly in RNA-binding proteins. The existence of an RRM-domain in L1ORF1p now explains why L1ORF1p binds LINE-1 RNA and how this could happen in detail. The insight into the structure of the L1ORF1p protein provides a new perspective and a good basis for future investigations of those cellular processes that are exploited by the LINE-1 element for its own propagation, and also for those mechanisms that are available to the cell to prevent the excessive propagation of retrotransposons.
Paper: Elena Khazina, Oliver Weichenrieder: Non-LTR retrotransposons encode noncanonical RRM domains in their first open-reading frame. PNAS, 20. Januar 2009, vol. 106 (3), 731-736, doi: 10.1073/pnas0809964106.
Provided by Max Planck Institute for Developmental Biology