Purifying parasites with light
Researchers have developed a clever method to purify parasitic organisms from their host cells, which will allow for more detailed proteomic studies and a deeper insight into the biology of organisms that cause millions of cases of disease each year.
Many infectious pathogens, like those that cause Toxoplasmosis or Leishmaniases, have a complex life cycle alternating between free-living creature and cell-enclosed parasite. A thorough analysis of the proteins that help these organisms undergo this lifestyle change would be tremendously useful for drug or vaccine development; however, it's extremely difficult to separate the parasites from their host cell for detailed study.
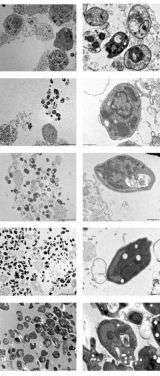
As reported in the September Molecular & Cellular Proteomics, Toni Aebischer and colleagues worked around this problem by designing special fluorescent Leishmania mexicana (one of the many Leishmaniases parasites). They then passed infected cells through a machine that can separate cell components based on how much they glow. Using this approach, the researchers separated the Leishmania parasites with only about 2% contamination, far better than current methods.
They then successfully identified 509 proteins in the parasites, 34 of which were more prominent in parasites than free –living Leishmania. The results yielded many characteristics of these organisms, such as a high presence of fatty acid degrading enzymes, which highlights adaptation to intracellularly available energy sources. The identified proteins should provide a good data set for continued selection of drug targets, and the success of this method should make it a good resource for other cellular parasites like malaria.
Citation: "Transgenic, Fluorescent Leishmania mexicana Allow Direct Analysis of the Proteome of Intracellular Amastigotes" by Daniel Paape, Christoph Lippuner, Monika Schmid, Renate Ackermann, Martin E. Barrios-Llerena, Ursula Zimny-Arndt, Volker Brinkmann, Benjamin Arndt, Klaus Peter Pleissner, Peter R. Jungblut, and Toni Aebischer
Article URL: www.mcponline.org/cgi/content/full/7/9/1688
Source: American Society for Biochemistry and Molecular Biology



















