GIANT-Coli: A novel method to quicken discovery of gene function
Think researchers know all there is to know about Escherichia coli, commonly known as E. coli? Think again. "E. coli has more than four thousand genes, and the functions of one-fourth of these remain unknown," says Dr. Deborah Siegele, a biology professor at Texas A&M University whose laboratory specializes in carrying out research using the bacterium.
Harmless E. coli strains are normally found in the intestines of many animals, including humans, but some strains can cause diseases.
Siegele and her co-workers at the University of California San Francisco, Nara Institute of Science Technology and Purdue University have devised a novel method that allows rapid and large-scale studies of the E. coli genes. The researchers believe their new method, described in the current online issue of Nature Methods, will allow them to gain a better understanding of the E. coli gene functions.
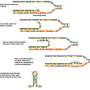
The principle behind this new method is genetic interaction. Interaction between genes produces observable effects, and this allows researchers to identify the gene functions. The research team has called their new method GIANT-Coli, short for genetic interaction analysis technology for E. coli.
The team believes that its method has great potential to quicken the progress of discovering new gene functions. The use of GIANT-Coli has already allowed researchers to identify some previously unknown genetic interactions in E. coli.
To study genetic interaction, researchers need to use what they call double-mutant strains. GIANT-Coli allows large-scale generation of these double-mutant strains (high-throughput generation). And this is the first time that a high-throughput generation method for double mutants of E. coli has been developed.
Why is it so important to know the E. coli better? "Much of what we know about other bacteria, including the more dangerous ones like Vibrio cholerae, comes from our knowledge of E. coli," says Siegele. "The E. coli is a model organism."
Siegele says that GIANT-Coli can be developed to study genetic interactions in other bacteria, and because some proteins are conserved from bacteria to humans, perhaps some of the results can even be extrapolated to gene function in humans. Moreover, Siegele points out that the method has obvious application in medicine because understanding gene functions in harmful bacteria will help in developing better treatment approaches.
Source: Texas A&M University