8 new human genome projects offer large-scale picture of genetic difference
A nationwide consortium led by the University of Washington in Seattle has completed the first sequence-based map of structural variations in the human genome, giving scientists an overall picture of the large-scale differences in DNA between individuals. The project gives researchers a guide for further research into these structural differences, which are believed to play an important role in human health and disease. The results appear in the May 1 issue of the journal Nature.
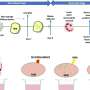
The project involved sequencing the genomes of eight people from a diverse set of ethnic backgrounds: four individuals of African descent, two of Asian descent, and two of European background. The researchers created what's called a clone map, taking multiple copies of each of the eight genomes and breaking them into numerous segments of about 40,000 base pairs, which they then fit back together based on the human reference genome. They searched for structural differences that ranged in size from a few thousand to a few million base pairs. Base pairs are one of the basic units of information on the human genome.
Most previous studies of the genome have focused on small genetic variations called SNPs (pronounced "snips"), or single-nucleotide polymorphisms -- changes on the scale of a single base pair. More recent research on the human genome has shown, however, that larger-scale differences may account for a great deal of genetic variation among individuals. Structural variation in the human genome has already been linked to individual differences in susceptibility to conditions like coronary heart disease, HIV, schizophrenia, autism, and mental retardation.
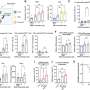
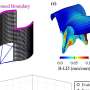
In addition to millions of smaller differences, the researchers identified 1695 regions of structural variation in the genome. They also provided a detailed look at the sequence for 261 regions of the genome, revealing an unprecedented view of the complexity of the genetic differences among different humans. The large-scale differences that the researchers were looking for can come in many forms, such as the deletion of a large swath of DNA, or the insertion of an out-of-place string of genetic code. Others simply appear as a different number of copies of a gene or DNA sequence.
Until now, there has not been a comprehensive study to sequence these variations systematically in multiple individuals. As part of their study, the authors also discovered 525 segments of DNA that were previously unknown to the human genetics community.
"There is a perception that the human genome is essentially completely understood," explained the project's leader, Dr. Evan Eichler, UW associate professor of genome sciences and an investigator for the Howard Hughes Medical Institute. "The sequences we have identified range in size from a few thousand to hundreds of thousands of base pairs, and are not part of the published human genome reference sequence. We found that many of these are highly variable in copy and content between individuals. This represents uncharted territory that can now be examined in more detail to determine the function of these new segments of the human genome with respect to disease and gene activity."
Eichler expects that the structural variation map will give scientists a much better picture of genetic variations, and help them better understand these areas of the genome that are prone to large-scale changes over time. Even more research is needed on structural variations, the scientists argue in the article, to help get a more accurate picture of the human genome than what we already have in the reference genome constructed by the Human Genome Project.
"The important point here is that we could not have found these differences without sequencing more human genomes from individuals of diverse ancestry to a high-quality standard," Eichler added.
The project will also serve as a sound resource for the science community, said Eichler, since the researchers have preserved the many segments of DNA used for the project. As new genomes are studied, someone might find a new sequence or new area of variation, and the researchers can revisit that particular segment of DNA to study it more closely.
Source: University of Washington