Tuatara evolving faster than any other species
New DNA research has questioned previous notions about the evolution of the tuatara.
In a study of New Zealand’s “living dinosaur” the tuatara, evolutionary biologist Professor David Lambert and a team from the Allan Wilson Centre for Molecular Ecology and Evolution have recovered DNA (deoxyribonucleic acid) sequences from the bones of ancient tuatara up to 8000 years old.
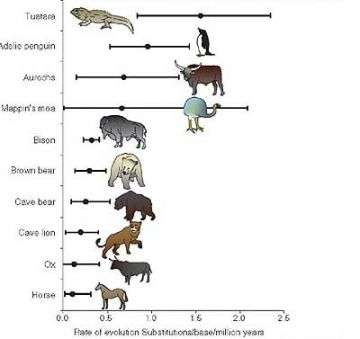
They found that although tuatara, have remained largely physically unchanged over very long periods of evolution, they are evolving - at a DNA level - faster than any other animal yet examined. The research has just been published in the prestigious international journal Trends in Genetics and features on the cover of the issue.
“What we found is that the tuatara has the highest molecular evolutionary rate that anyone has measured,” Professor Lambert says.
The rate of evolution for Adélie penguins, which Professor Lambert and his team have studied in the Antarctic for many years, is slightly slower than that of the tuatara. The tuatara rate is significantly faster than for animals including the cave bear, lion, ox and horse.
“Of course we would have expected that the tuatara, which does everything slowly – they grow slowly, reproduce slowly and have a very slow metabolism – would have evolved slowly. In fact, at the DNA level, they evolve extremely quickly, which supports a hypothesis proposed by the evolutionary biologist Allan Wilson, who suggested that the rate of molecular evolution was uncoupled from the rate of morphological evolution.”
Allan Wilson, who died of leukaemia in 1991, was a pioneer of molecular evolution. His ideas were controversial when introduced 40 years ago, but this new research supports them.
Professor Lambert says the finding will be helpful in terms of future study and conservation of the tuatara, and the team now hopes to extend the work to look at the evolution of other animal species.
“We want to go on and measure the rate of molecular evolution for humans, as well as doing more work with moa and Antarctic fish, to see if rates of DNA change are uncoupled in these species. There are human mummies in the Andes and some very good samples in Siberia where we have some collaborators, so we are hopeful we will be able to measure the rate of human evolution in these too.”
The tuatara, Sphendon punctatus, is found only in New Zealand and is the only surviving member of a distinct reptilian order, Sphehodontia, that lived alongside early dinosaurs and separated from other reptiles 200 million years ago in the Upper Triassic period.
Source: Massey University