CSHL researchers discover three new genes that cause lung cancer
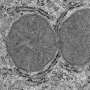
Researchers at Cold Spring Harbor Laboratory (CSHL) have discovered three genes that interact with cancerous results in 20% of lung cancers. The three genes are located next to each other on human chromosome 14 and two are known to play key roles in fetal lung development.
According to CSHL lead investigator David Mu, “lung cancer cells in adults can reactivate genes that are normally active in the earliest stages of lung development. We identified the mutation that triggers this abnormal re-activation of developmental genes and showed that if you turn off these genes, you stop the cancer.”
The CSHL research found that the three genes termed TTF1, NKX2-8, and PAX9 interact to reactivate what appears to be an early fetal gene expression pattern that results in cancer tumor growth. “The collaboration of these genes and the fact that they are so close together on the chromosome may explain why this mutation is so common in lung cancer,” said CSHL investigator and co-author Scott Powers. In collaboration with Dr. William Gerald at the Memorial Sloan Kettering Cancer Center, the study finds that the mutation is more prevalent in late stage lung cancer and is possibly a risk factor for recurrence.
The CSHL-led research demonstrates that the cancerous results of the mutation can be reversed. In the future, this may lead to new treatment options for patients. Cancer research that looks at one gene at a time ignores the fact that cancers are usually caused by multiple collaborating cancer genes. Mutations in these genes determine the clinical outcome of the cancerous growth and how the cancer responds to treatment.
“At CSHL we are excited about the ability to apply direct genomic analysis to human cancers and discover more about how cancer genes interact,” said Howard Hughes Medical Institute Investigator and CSHL Cancer Center Deputy Director Scott Lowe.
Source: Cold Spring Harbor Laboratory