Gene regulation in humans is closer than expected to simple organisms
Using a novel method developed to identify reliably functional binding motifs, researchers from the Weizmann Institute of Science in Israel have performed a genome-wide study of functional human transcription factor binding sites that encompasses nearly ten thousand genes and four hundred known binding motifs. The study appears in the Aug. 29 issue of the online, open-access journal PLoS ONE.
Gene networks are some of the most basic features of a living organism. An external or internal stimulus activates some genes, which in turn control others genes whose activity turns on or off various biological processes (such as the cell cycle, energy production, DNA repair, cellular suicide etc).
Many of the regulatory functions are controlled by attachment of special proteins (transcription factors) to 6 - 10 nucleotide long binding sequences located on the DNA, activating or suppressing expression of the regulated gene. Our ability to identify these binding sites is essential to understand the way biological networks operate.
As the genomes of various organisms became known, it turned out that complex and simple organisms differ less than anticipated in the sizes and makeup of their genomes; complexity of an organism is now believed to be reflected mainly in the manner in which expression is regulated. According to consensus, transcription of human genes is regulated predominantly by factors that bind to sites whose distances from the transcription start site may vary widely and reach tens of thousands of base pairs.
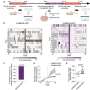
To test the validity of this belief/consensus, researchers from the Weizmann Institute of Science in Israel have performed a genome-wide study of functional human transcription factor binding sites that encompasses nearly ten thousand genes and four hundred known binding motifs. Using a novel method that was developed to identify reliably functional binding motifs, they discovered that in human (and mouse) a surprisingly large fraction of the functional binding sites was concentrated very close to the transcription start site. Hence on the basis of currently available data it seems that the most basic underlying principles and strategies used by the genomes of higher organisms to regulate gene expression are quite close to those used by simple organisms like bacteria and yeast.
The discovery and the method will allow more focused and reliable search for transcriptional binding sites and hence may turn into a major tool to be used in the quest for the transcriptional networks whose function governs all cellular processes, and whose breakdown causes complex diseases. It will generate progress in establishing the design principles used by the transcription process in high organism, and allow a more focused search for the origins of their complexity.
Source: Public Library of Science