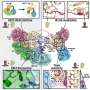
Computer simulation shows how evolution may have speeded up
Is heading straight for a goal the quickest way there? If the name of the game is evolution, suggests new research at the Weizmann Institute of Science, the pace might speed up if the goals themselves change continuously.
Nadav Kashtan, Elad Noor and Prof. Uri Alon of the Institute’s Molecular Cell Biology and Physics of Complex Systems Departments create computer simulations that mimic natural evolution, allowing them to investigate processes that, in nature, take place over millions of years.
In these simulations, a population of digital genomes evolves over time towards a given goal: to maximize fitness under certain conditions. Like living organisms, genomes that are better adapted to their environment may survive to the next generation or reproduce more prolifically. But such computer simulations, though sophisticated, don’t yet have all the answers. Achieving even simple goals may take thousands of generations, raising the question of whether the three-or-so billion years since life first appeared on the planet is long enough to evolve the diversity and complexity that exist today,
Evolution takes place under changing environmental conditions, forcing organisms to continually readapt. Intuitively, this would slow things down even further, as successive generations must switch tack again and again in the struggle to survive. But when Kashtan, Noor and Alon created a simulation in which the goals changed repeatedly, they found that its evolution actually speeded up. They even found that the more complex the goal – i.e., the more generations needed reach it under fixed conditions – the faster evolution accelerated in response to changes in that goal.
Computerized evolution ran fastest, the scientists found, when the changes followed a pattern they believe may be pervasive in nature. In previous research, Kashtan and Alon had shown that evolution may often be modular – involving adjustments to standard parts, rather than wholesale remodeling. They theorized that the forces acting on evolution may be modular as well, and for each goal, they defined subgoals that could each change in relation to the others. 'In an organism, for example, you might classify these subgoals as the need to eat, the need to keep from being eaten, and the need to reproduce. The same subgoals must be fulfilled in each new environment, but there are differences in nuance and combination,' says Kashtan. 'We saw a large speedup, for instance, when we repeatedly exchanged an 'OR' for an 'AND' in the computer code defining our goals, thus changing the relationship between subgoals.'
Although the main aim of this research, which appeared recently in the Proceedings of the National Academy of Sciences, was to shed light on theoretical questions of evolution, it may have some practical implications, particularly in engineering fields in which evolutionary tools are commonly used for systems design; and in computer science, by providing a possible way to accelerate optimization algorithms.
Source: Weizmann Institute of Science