Scientists decode RNA mystery, will help aim drug therapies
A team of University of Maryland scientists have made a discovery that will help better direct drug therapies to their molecular targets.
As reported in the June 13 issue of the online journal PLoS ONE, the researchers, led by Jonathan Dinman, assistant professor of cell biology and molecular genetics at the University of Maryland, have found the difference between two closely related components in the messenger RNA (mRNA) – near-cognate and non-cognate codons – terms that have long been used, but not understood.
“Although these two terms have been used by scientists for over 40 years, the differences between them have never been properly defined. Here, we have made this determination at both the molecular and mechanistic levels, and developed a simple drug-based test to differentiate them. It’s a real step in designing the pathway of rational drug design.”
Defining the Codons
Messenger RNA tells the ribosome what kind of proteins to make and how much to crank out. “The codons specify which amino acids should end up in the protein,” says Dinman. “If the wrong codon is selected, the wrong amino acid ends up in the protein, which can alter or destroy the function of that protein and cause human disease.”
Codons are the three-part packets of information in DNA that specify amino acids. They are read by corresponding packets in aminoacyl tRNAs (aa-tRNA) called anticodons. Codons are among the many components that James Watson, Francis Crick, François Jacob, Jacques Monod, Marshall Nirenberg and others found in their discoveries of DNA and genetic code in the late 1950’s and early 1960s.
“Crick coined the terms ‘near-cognate’ and ‘non-cognate codons’ but he didn’t really define the difference,” says Dinman. “In fact, no one really ever has defined them, until now. It’s been fuzzy.”
“Cognate tRNAs properly decode genetic information in mRNAs, while near- and non-cognate tRNAs correspond to ‘close but no cigar’ and ‘fuggedaboutit,’ respectively,” says Dinman. “Although ribosomes mistakenly misinterpret both near- and non-cognate codons very infrequently (about a one in five thousand chance), exactly how they might be misread is different.”
Hitting the Target
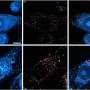
Using firefly luciferase, the protein that makes fireflies glow, Dinman’s team discovered that the difference between near- and non-cognate aa-tRNAs lies in the potential, albeit imperfect, of near-cognate aa-tRNAs to pair up with messenger RNA at all three positions to create three sets of base pairs. This serves as the signal to tell the ribosome to use that aa-tRNA. In contrast, that potential can never exist between mRNAs and non-cognate aa-tRNAs.
“There are drugs that can fool the ribosome into misreading aa-tRNAs, thus providing a way to suppress or bypass a mutation in a gene” says Dinman. “ However, some drugs work one way and may be best suited for making ribosomes use near-cognate aa-tRNAs, while others work another way, and are best used for non-cognate cases. The nature of the mutation responsible for a specific disease, whether it is near- or non-cognate, will determine the therapeutic strategy.”
By defining these differences in the codons, Dinman team’s findings will help researchers improve their accuracy in designing drug therapies that suppress mutations. “You need to know whether you’re targeting the near or non-cognate codon to get the right message to the ribosome,” Dinman says. “The clearer the view of the target, the easier it is to hit.”
Source: Public Library of Science