A new method for mapping gene expression deep in the body could one day be used to track stem cells or cancer therapy
Even fans of black-and-white film can't deny that color brought new life to photography and motion pictures. And when it comes to learning what happens inside the body, there's no substitute for color. Were it possible, for example, to map out real-time gene expression in our body's cells using contrasting colors, scientists would gain a glimpse of vital biological processes that are currently invisible. The problem is that the glowing multicolor proteins used by scientists to illuminate the inner workings of cells are of little help in observing deep-seated processes in the body, because the thickness of tissues obscures the glow.
Researchers at the Weizmann Institute of Science have now demonstrated the possibility of observing such processes by means of magnetic resonance imaging, or MRI, whose radio-wave signals—as opposed to light's glow—are not stopped by tissues, no matter how thick. As reported today in Nature Biotechnology, the researchers have developed a method for using MRI to simultaneously track, in two colors, the expression of two different genes. The method paves the way for using MRI to observe a wide range of biological processes within the living body in research and in the clinic. When developed further, it may serve, for example, to examine how one brain region affects another, monitor the effects of cancer therapy or trace the fate of stem cells introduced into the body for therapeutic purposes.
"MRI may one day be used to peer deep into the body over an extended period of time, to see what goes on in tissues without the need to remove them for study under a microscope," says Dr. Amnon Bar-Shir of the Molecular Chemistry and Materials Science Department, who headed the research team. "Our method provides a major step in that direction."
When the Nobel Prize in Chemistry was awarded in 2008 for the development of fluorescent proteins to be used as "reporters" for viewing gene expression under the microscope, one of the laureates, the late Roger Y. Tsien, stated in his acceptance speech that such fluorescence has its limitations, and that in the future these might be overcome by techniques such as MRI. That was a visionary statement, yet the familiar grayscale MRI scans that are used for diagnosis do not display gene activity, but only structural elements. Nor are the advanced multicolor MRI images, generated under certain circumstances, suitable for revealing gene expression. When MRI was in the past adapted to reflect gene expression, it could "report" on only one gene at a time, by picking up a change in the strength of the signal, which is recorded as a dark dot in the black-and-white image. The crucial missing element needed for monitoring biological processes was the ability to detect the expression of several genes simultaneously. As in fluorescent labeling, this would ideally involve assigning different colors to the genes.
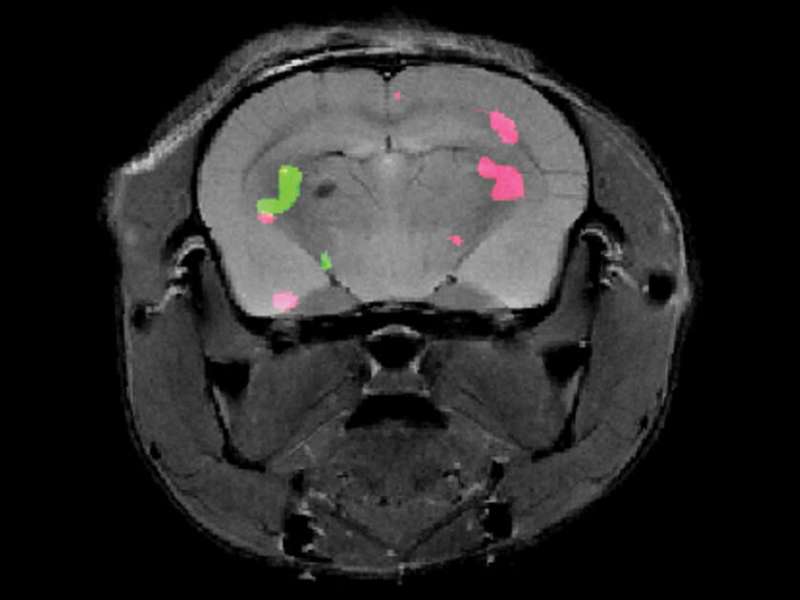
To address the challenge, the Weizmann scientists developed a two-step method: First, they genetically engineered two groups of cells, so that each group expressed one of two specially designed proteins. In parallel, the scientists created a mixture of two types of molecular probes, intended to be injected into the bloodstream and to accumulate exclusively in the cells expressing the engineered proteins. The two probes had been designed to issue a signal in response to different MRI frequencies, each lighting up in a different color.
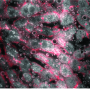
Dr. Hyla Allouche-Arnon, a staff scientist in Bar-Shir's lab, led the study, in which Bar-Shir's team and other researchers collaborated with Prof. Sarel Fleishman and Dr. Olga Khersonsky of the Biomolecular Sciences Department. Applying the method in live mice, the scientists used exceptionally powerful MRI equipment with a magnet of about 15 tesla—one of only a handful of such machines in the world. The scans picked up the frequencies of the molecular probes, revealing the exact positions of the cells that expressed each of the proteins and marking them in green and pink.
"Gene expression lets us know what each cell is doing," Allouche-Arnon says. "Thanks to our method, MRI may now be applied by researchers in various fields to track the activity of all kinds of processes, for example, those involving different types of brain or immune cells."
The approach may be further developed to simultaneously map, in color, the expression of more than two genes. And if adapted for use in humans, it may enable researchers and physicians to watch important processes in action in a noninvasive manner. For example, in cell therapy for cancer, different color probes could be used to track the relative positions of the tumor and the therapeutic cells.
Study participants also included Nishanth D. Tirukoti, a research student in Bar-Shir's lab; Dr. Yoav Peleg, Dr. Orly Dym, Dr. Shira Albeck, Dr. Alexander Brandis and Tevie Mehlman of Weizmann's Life Sciences Core Facilities Department; Dr. Liat Avram and Dr. Talia Harris of Weizmann's Chemical Research Support Department; and Dr. Nirbhay N. Yadav of the Johns Hopkins School of Medicine.
More information: Hyla Allouche-Arnon et al, Computationally designed dual-color MRI reporters for noninvasive imaging of transgene expression, Nature Biotechnology (2022). DOI: 10.1038/s41587-021-01162-5
Journal information: Nature Biotechnology
Provided by Weizmann Institute of Science