High-fidelity recording of molecular geometry with DNA 'nanoscopy'

Researchers are constantly expanding their arsenal of methods to decipher the spatial organization of biological structures. Using microscopes, they can now visualize individual macromolecular components within DNA, protein, or other complexes. However, this resolution typically requires sophisticated equipment applied to specially-processed samples, and it is difficult to simultaneously watch many types of molecules, especially at high density and throughput, or dynamic interactions.
Circumventing the need for expensive microscopes, some recent biochemical approaches attach barcoded DNA probes to molecular targets and then fuse those in nearby pairs together, often by DNA ligation. These DNA "records" are later read out for analysis. Because these methods destroy the DNA probes in the process of pairing, however, the information acquired from each molecular target cannot include more than one interaction, neither multiple at once nor one changing over time. Such methods can severely limit the quality of any subsequent computational reconstruction, and make reconstruction of individual complexes impossible.
To overcome these limitations, a team at Harvard's Wyss Institute of Biologically Inspired Engineering led by Core Faculty member Peng Yin, Ph.D., has now developed a DNA nanotechnology-based method that allows for repeated, non-destructive recording of uniquely barcoded molecular pairings, rendering a detailed view of their components and geometries. In the future, the approach could help researchers understand how changes in molecular complexes control biological processes in living cells. The study is published in Nature Communications.
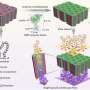
"Our method, which we call "Auto-cycling Proximity Recording" (APR), essentially acts as a continuous biochemical recorder of the molecular structures," said Yin, who is also Professor of Systems Biology at Harvard Medical School. "APR allows us to look at many proximities simultaneously and repeatedly, and with minimal distrurbance to structure. By assessing the full complement of all such pairs in many cycles, we can create a detailed view of a molecular structure and even observe different structural states of the same targets."
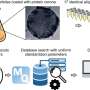
As proof-of-principle, the team designed multiple DNA probes in silico, and synthesized and attached them to molecular targets contained in the prescribed geometries of DNA origami nanostructures. Through this newly-engineered, DNA-directed biochemical mechanism, a record in the form of a barcoded DNA strand is synthesized on the structure if and only if two of these DNA probes are in close enough proximity to each other ("proximity recording"). Records are released as they are synthesized, and later collected for sequence analysis.
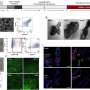
Unlike other biochemical methods, each individual APR target can yield over 30 DNA records ("auto-cycling"), allowing robust data collection. After collecting all DNA records, the team compiled their sequences and successfully reconstructed the geometry of the synthetic nanostructures. Thus, the approach functions as a 'DNA nanoscope', which uses specifically engineered DNA biochemistry to visualize target pairs in a molecular object. Expanding on these new capabilities, the Wyss researchers were even able to document changes in the state of individual nanostructures, raising the possibility that the approach could be used to correlate structural transitions in molecular complexes with their biological functions.
"By using antibodies and other widely used agents to direct DNA probes to molecular targets, we could apply APR technology to decode the components and geometries of biological complexes," said Thomas Schaus, M.D., Ph.D., a Wyss Institute Staff Scientist who as the study's first author together with Yin developed APR. "The fact that individual DNA records carry unique, sequenceable barcodes and that the method is scalable may enable us to one day follow, individually, thousands or millions of macromolecules in a biochemical pathway."
"The development of APR as a nanotechnological means to decipher molecular structures without the need for elaborate and expensive microscopes really illustrates how the Wyss Institute's recently launched Molecular Robotics initiative can impact structural biology research and developments in many laboratories," said Wyss Institute Founding Director Donald Ingber, M.D., Ph.D., who also is the Judah Folkman Professor of Vascular Biology at HMS and the Vascular Biology Program at Boston Children's Hospital, as well as Professor of Bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences.
Journal information: Nature Communications
Provided by Harvard University