Image-based modeling of body tissues
A team of researchers from the New Jersey Institute of Technology (NJIT), in collaboration with their colleagues from the University of Oklahoma, have demonstrated a novel, image-based simulation approach that could lead to artificially grown tissues and organs in a hospital setting.
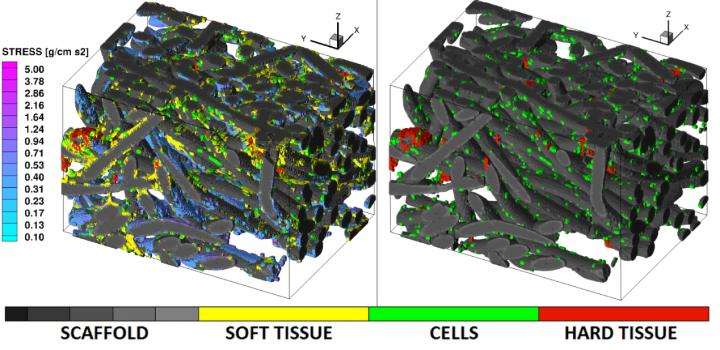
Combining bone tissue engineering (BTE) experiments, micro-computed tomography (uCT) sample scanning, "virtual histology" image segmentation, and lattice Boltzmann method (LBM) fluid dynamics, the researchers produced realistic simulations of BTE scaffolds cultured in flow perfusion bioreactors. Understanding the interplay between scaffold manufacturing parameters, culturing conditions and cell biology within the construct is necessary for transitioning regenerative medicine to a clinical setting. Although previous attempts have been made at modeling artificial tissue cultures, they were limited by oversimplifying assumptions, such as a uniform cell / tissue monolayer coverage of the scaffold's surface and idealized scaffold geometries.
This novel and scalable technology enables researchers to account for the realistic architectural non-idealities inherent to tissue engineering scaffolds and the presence of cells / tissues in their pores. An additional advantage of this method is that it allows for correlating cell behavior and tissue growth with the flow physics occurring inside of complex 3-D scaffold microenvironments. Moreover, such relationships can be tracked over time via modeling based on nondestructive repeated scanning. The report appears in the December 2016 issue of the journal Technology.
"By taking advantage of the exact spatio-temporal information provided by the high-resolution micro-CT imaging, this approach opens the door for transforming computer-assisted tissue engineering, which is traditionally done based on virtual drawings of scaffolds and very little or no cross-validation against experiment," says Professor Roman Voronov, Ph.D. of the New Jersey Institute of Technology and principal investigator of the paper.
The manuscript offers a 'recipe' for the technology that culminated from a decade-long effort by the team working on the BTE-modeling problem. While the imaging aspect of their approach offers an unprecedented ability to detect and distinguish individual cells, soft tissues and calcification embedded within the scaffold, the LBM has the ability to handle large-scale simulations (such as those resulting from the sub-micron voxel resolution shown here) and complex boundary conditions typical of the BTE scaffolds. And although the results are only meant to serve as proof of concept, the demonstrated technology is not limited by sample size, since its algorithms are fully parallelizable for supercomputing.
Therefore, it is straightforward to extend to full-scaffold models, with the only physical limitation being the micro-CT sample-chamber. However, those are typically much larger than the scaffolds.
"Moreover, in case repeated scanning is not possible, or not desirable, the number of scans required can be minimized by simply subtracting any bio-matter from an end-point image of fully cultured scaffolds. This would help to estimate the scaffold's initial geometry prior to the cell seeding," said Taseen Alam of NJIT, the first author of the paper. Once this is done, the fluid flow patterns established within the initial empty scaffold can be correlated to the tissue growth observed in the end-point image. Finally, since the calculated results are overlaid onto the experimental images in 3-D, the method itself serves as a direct comparison to the experiment. And any correlation obtained as a result of the image-based modeling can be subsequently tested by attempting predictions in samples previously unencountered by the code.
The team is now working to extend the technology by including molecular transport, which is rarely simulated by the conventional models. For example, distribution of O2 and nutrient/waste within the scaffold, transport of scaffold degradation byproducts whose acidic nature may be detrimental to the cells, and molecular signals between the cells could all be accounted for using the same image-based approach. In this way, a more complete picture of cell behavior in complex micro-environments can be generated.
More information: Taseen A. Alam et al, Image-based modeling: A novel tool for realistic simulations of artificial bone cultures, Technology (2016). DOI: 10.1142/S233954781620003X
Provided by World Scientific Publishing