The process of DNA packaging in cell nucleus revealed
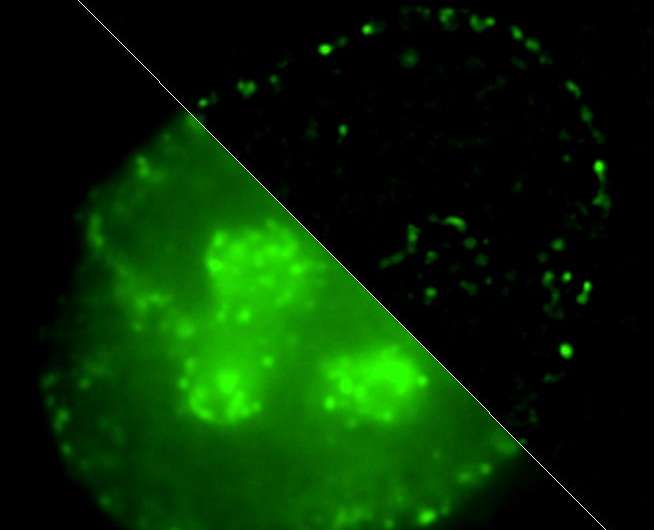
Scientists from the Lomonosov Moscow State University in collaboration with their colleagues from the U.S. have conducted a study on DNA packaging in the cell nucleus and its alteration in the process of DNA replication. DNA packaging is supposed to have strong influence on genes' activity. It's also one of the mechanisms of epigenetic control of gene expression.
Biologist Igor Kireev, the chief of the Electron Microscopy Department at the Belozersky Institute of Physico-Chemical Biology and one of the article authors, says, "Although all the cells in a given organism possess similar genetic information, not all genes operate in a particular cell type. In fact, the set of active genes determines the cell fate. There are some cellular mechanisms assisting the cell to remember which genes should be activated and which should not. This is the epigenetic control or, in other words, cell memory, that determines what the cell is, and the genes that directed it to form in such a way. We were interested in so-called high levels of chromosome structural organization, which are formed through a series of consecutive phases in DNA packaging."
DNA exists in a cell as a complex of proteins—chromatin. Initial stages of chromatin compaction are studied quite well. These are nucleosomes, protein globules, each is around 10 nanometers long and consisting of 8 histone protein molecules, around which DNA is wrapped. Then a chain of nucleosomes is packed in a way that we don't know yet, resulting in thicker fibrils and chromonemata. At the end we get a very high degree of compaction. So, the length of a most densely compacted mitotic chromosome is 20 000 times smaller than the length of DNA, which is packed inside it.
Replication is the process of copying DNA molecule and transcription is the process of RNA synthesis, both using DNA as a template. It was traditionally assumed that high compaction of DNA in chromatin hinders these template-based synthesis processes and should be disassembled on a scale of quite large chromatin domains. It was fairly difficult to identify these domains and analyze their structural organization at high spatial resolution without disturbing their natural structure.
Igor Kireev comments: "We've offered a method of labeling chromatin domains based on differences in replication timing of active and inactive gene loci. While actively transcribed chromosome regions—so called euchromatin— replicate at the very beginning of synthetic phase of the cell cycle, inactive 'silent' heterochromatin replicates during its second half. Our technique allows nondestructive detection of these two fractions of chromatin at the electron microscopy level. It's based on a combination of new synthesized DNA labeling using click chemistry methods with subsequent detection of reaction products by correlative super-resolution optical microscopy and immuno-electron microscopy. In other words, we can see one and the same molecule in this cell both in optical and electron microscopes."
Application of this approach has yielded two surprising observations. First of all, "working" chromatin, contrary to traditional ideas, maintains a quite high level of packaging, as it's represented by highly structured chromatin fibrils of higher order—chromonemata. Secondly, it turns out that DNA in the chromonema structure possesses a high degree of structural flexibility, which means that it's able to flow from one section of chromonema fiber to the neighboring one. However, the general dense structure of the chromonema doesn't change. These observations suggest new hypotheses concerning mechanisms of epigenetic information transfer in the process of cell division.
Igor Kireev says, "We've made an assumption that newly synthetized DNA in chromatin structures isn't fixed, but could move inside them, interacting with DNA segments that haven't doubled yet, and which 'remember everything.' Moreover, this DNA contains the necessary molecular components for recovery of lost epigenetic information."
Another result mentioned in the article, which has been published in Current Biology, is that genome structural organization doesn't have strict hierarchy. There are, of course, some gradual levels of DNA organization. It was thought earlier that there should be a strictly ordered system, which uniquely leads to chromosome formation. But now, it's turned out that everything could be organized in a different way—there are some building principles, but within specified chromatin frameworks, DNA has some freedom and flexibility.
Igor Kireev shares future plans: "Further research development includes, first of all, transition to analysis of individual chromosome loci. We are going to label them both on optical and electron microscopic levels with the help of TALE technology. And secondly, further research will address elaboration of even more native methods of in vivo labeling that are compatible with such advanced technologies as cryo-electron microscopy."
Scientists hope to observe DNA spatial organization in detail, applying direct methods of visualization of chromatin packaging methods with high resolution. The research will help to clarify structural aspects of epigenetic control of gene expression, and possibly offer some insight into its regulation. This will contribute to elaboration of more effective approaches to therapeutic intervention—for instance, to fight cancer or aging, where the epigenetic component plays an important role.
More information: Xiang Deng et al, Cytology of DNA Replication Reveals Dynamic Plasticity of Large-Scale Chromatin Fibers, Current Biology (2016). DOI: 10.1016/j.cub.2016.07.020
Journal information: Current Biology
Provided by Lomonosov Moscow State University