Shining light on disease
Cyanine dyes could improve the efficiency of molecular probes in identifying, for example, the presence of a virus or a tumor receptor.
Scientists use fragments of RNA and DNA with specific nucleotide sequences to identify others with complementary sequences, indicating, for example, the presence of a specific kind of virus.
Researchers in Japan have recently improved the existing probing techniques using a cyanine dye called Cy3.
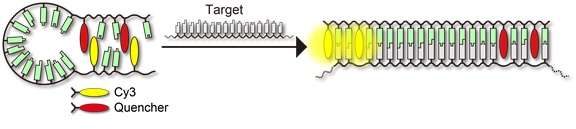
RNA and DNA "probes" are conventionally made using fragments of strands that have nucleotide sequences on either end that complement each other. A fluorescent chemical compound, called a "fluorophore", is added to one end of the probe and a "quencher" is added at the other. In the absence of its complementary "target", the single-stranded probe comes together in a hair-pin-like manner, with the complementary sequences at either end of the strand binding together, bringing the fluorophore and quencher close to each other and turning down, or "quenching", the fluorophore's fluorescence. However, when a probe is present in a sample with its complementary target RNA, the probe strand "opens up" to combine with its target, allowing its detection when light is shone and the fluorophore fluoresces.
This method is widely used, but the response of the hair-pin-like probe to its targets is relatively slow.
A team of researchers from Nagoya University and the Japan Science and Technology Agency developed a new probe design based on Cy3 as the fluorophore. Cy3 and the quencher (nitro methyl red) were incorporated into either end of a linear strand that lacked the self-complementary sequences found in conventional probes. Even so, Cy3 and the quencher were drawn to each other spontaneously to form a highly stable complex in the absence of a target, quenching Cy3's fluorescence.
In the presence of a complementary target, a strong fluorescence response was observed, which was ten times faster compared to conventional techniques.
The team optimized its design by incorporating two Cy3 residues separated by two nucleotide bases on one end of the probe strand, and two nitro methyl red residues separated by two nucleotide bases on the other end. This optimized design detected RNA with high efficiency and sensitivity.
Because Cy3 and nitro methyl red are able to combine in the probe, quenching fluorescence, without the need for self-complementary pairing, "this strategy will be applicable to the design of peptide-based probes," conclude the researchers in their paper published in the journal Science and Technology of Advanced Materials. Peptide probes are chains of linked amino acids that can be designed to bind with specific cell receptors, making them useful in tumor receptor imaging, for example.
More information: Hiromu Kashida et al. A stem-less probe using spontaneous pairing between Cy3 and quencher for RNA detection, Science and Technology of Advanced Materials (2016). DOI: 10.1080/14686996.2016.1182412
Journal information: Science and Technology of Advanced Materials
Provided by National Institute for Materials Science