November 29, 2016 feature
Managing complexity: Novel protein folding tool vastly simplifies understanding how sequence encodes structure
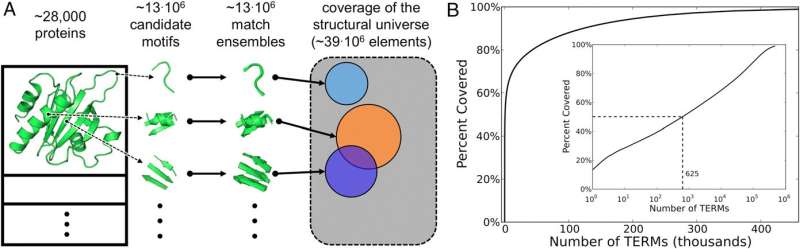
(Phys.org)—Protein folding is the process by which a polypeptide (a linear organic polymer chain consisting of many amino acid residues, or monomers) transforms from a random coil into the 3D conformation in which it can perform its biological function. Since different proteins fold into a range of very different shapes, the Protein Data Bank (PDB) – a database archive comprising experimentally-determined three-dimensional structures of large biological molecules, including numerous protein conformations – can be disarmingly complex. This is problematic because that space is fundamental to understanding how sequence encodes structure. Recently, however, scientists at Dartmouth College deconstructed the universe of known protein structures into reusable building blocks that they term tertiary structural motifs, or TERMs. (Structural motifs are compact blocks of a 3D protein structure.) They found that 50% of PDB protein conformations were described – at sub-Angstrom resolution – by a surprisingly small group of roughly 600 TERMs. Moreover, TERMs allowed them to discern sequence–structure relationships. The researchers state that these results can be used for protein structure prediction, protein design and other applications.
Prof. Gevorg Grigoryan discussed the paper that he and his co-authors published in Proceedings of the National Academy of Science of the United States of America. One of the primary challenges in their study was decomposing the set of known protein structures into standard reusable tertiary structural motifs. "The main challenge here was probably knowing where to begin," Grigoryan tells Phys.org. "Our overarching goal was to describe, in his words, an alphabet of protein structure." However, he points out that – unlike with text – the researchers were not able to visually determine where one structural unit (metaphorically a letter) began and another ended. "The number of ways in which we can potentially partition protein structure is extremely large, and so the task of finding a good decomposition seemed overwhelming."
The scientists addressed this problem by not defining a priori what the letters of the structural alphabet should be, but rather defining the task that these letters should accomplish – that is, describing the set of all residues and residue pair contacts observed in known protein structures. Next, they selected the smallest set of reusable building blocks they're named tertiary structural motifs, or TERMs, that would achieve this goal.
Another hurdle in determining the set of universal TERMs that capture all structure in the PDB was the difficult task of sifting through 13 million candidate TERMs, and describing which residues and contacts in known protein structures they individually explained. "Our previously-developed, efficient structure search algorithm MASTER helped us resolve this – but the total amount of computational time involved was still quite large, so we had to make use of a computer cluster." A computer cluster is a single logical unit comprising multiple networked-linked computers.

Grigoryan adds that by using residues and contacts rather than an a priori structural alphabet, defining the motif candidates was much easier. "It seemed particularly natural to define one candidate motif for every residue in the structural database," he notes, "such that the motif would capture the residue and all of its contacts – that is, the motif would describe that residue's local structural environment."
A key finding discussed in the paper was that universal TERMs provide an effective mapping between sequence and structure. "Because universal TERMs recur many times in unrelated proteins," Grigoryan tells Phys.org, "compiling the list of occurrences of each TERM allows us to start gleaning sequence rules that may underlie each of these structural motifs. The question was whether these sequence rules reflected fundamental determinants of structure, or simply noise from a limited structural database potentially biased by arbitrary evolutionary choices or the selection of proteins whose structures have been solved." The team resolved this through a series of experiments in which they demonstrated that a significant component of the sequence statistics emerging from TERM matches does likely emerge from fundamental sequence-structure relationships.
In effect, the natural utilization of TERMs provides a means of uncovering sequence–structure relationships. "Let's say a given TERM is consistent of a two-strand beta sheet interacting with an alpha helix at a particular characteristic crossing angle and distance," Grigoryan illustrates. "If we happen to have, for example, 600 instances of this motif from unrelated proteins, we essentially have 600 different examples of nature having made this structure with different amino-acid sequences. We can then use these 600 sequences to begin to understand what sequence features may be required or preferred to form such a structure - and we can do this for any TERM with sufficiently high usage in nature."
It turns out that by using this approach systematically for all TERMs contained in a given protein backbone structure, sequence variability predicted from TERM data agrees closely with evolutionary variation. "We can deduce a statistical model of what sorts of sequences would be likely to fold to that structure," he explains. "If we then ask this model to produce a whole bunch of such sequences, we find that the emergent sequence variability is often in close agreement to the evolutionary variability observe for the corresponding protein."
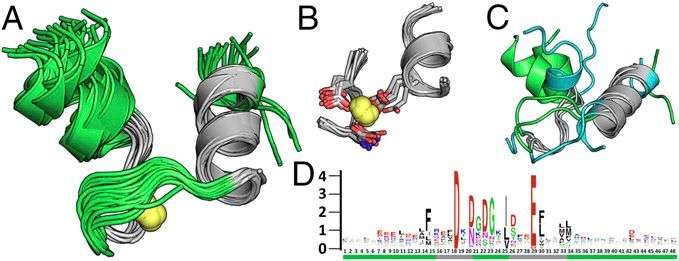
In addition, some 600 TERMs describe 50% of the known protein structural universe at sub-Angstrom resolution. "This refers to the level of degeneracy we discovered in the protein structure space." That only ~600 TERMs are required to describe half of all residues and inter-residue contacts in known protein structures suggests that at the local structural level, there just are not that many structural patterns that naturally emerge. There are, of course, a large number of more rare geometries, and full coverage of the protein structural universe requires tens or even hundreds of thousands of TERMs – but nevertheless, the majority of protein structure does appear to be quite degenerate at the local level."
As to the specific implications of their study for protein structure prediction, protein design, and other applications, Grigoryan points out that the major implication for protein design and structure prediction is the novel means of mining for sequence-structure relationships. "Statistical potentials, derived from known protein structures, have been employed for decades in both of these applications. However, such potentials typically describe the statistics associated with isolated simplistic structural features, like dihedral angles, individual interatomic or interresidue distances, or burial environments. However, TERMs offer the potential to describe sequence statistics in the context of holistic structural environments, which would be much more useful for both design and prediction." Specifically, he explains, in design, this would allow for a better understanding of what sequences would or would not form the target structure; for prediction, it would help drive structural sampling towards structures whose TERMs are most consistent with the modeled structure. "A potential limitation is the amount of available data, because not all TERMs have sufficient known instances to synthesize accurate sequence models," he acknowledges. "However, the early results shown in our paper, as well as some unpublished results in our lab, point to the fact that TERM-based statistics are already providing non-trivial insights that in many cases, other methods are unable to easily capture – and this is only going to get better as the amount of structural data continues to accumulate."
When asked about the implications of their work for synthetic genomics and synthetic proteomics, Grigoryan said "It's a good question. I can certainly see a future in which a truly robust method for computational protein design serves as a key element in synthetic genomics and proteomics applications. I'd say that in terms of our current design techniques, we're not quite there today - but our goal with TERM-based and other developments – as well as the general goal of our field – is certainly to keep improving the robustness of our methods, so that one day, we can offer them as black-box solutions to folks in other disciplines, whether that be materials science, biomedicine, or synthetic biology."
Moving forward, Grigoryan says that the team is focused on extending the capabilities of their TERM-based techniques to both protein design and structure prediction. "We're also very interested in introducing ensemble-based modeling approaches into protein design: Since protein structural states are really conformational ensembles, the language of statistical mechanics is most appropriate for describing their behavior. Therefore, we're pursuing methods for introducing statistical mechanics-based calculations towards improving the accuracy and robustness of protein design methods."
In addition to protein design and structure prediction, Grigoryan sees their study as having strong implications for our fundamental understanding of protein structure in general. "I think the new look at the protein structural universe our study offers can help not only with modeling and designing proteins, but it can also help with teaching about protein structure. The ideas of modularity and representation of standard motifs," he concludes, "have already made their way into my own teaching here at Dartmouth."
More information: Tertiary alphabet for the observable protein structural universe, PNAS November 3, 2016, Published online before print, doi:10.1073/pnas.1607178113
Journal information: Proceedings of the National Academy of Sciences
© 2016 Phys.org