New text-mining tool lets researchers visualize gene, protein, drug and disease connections
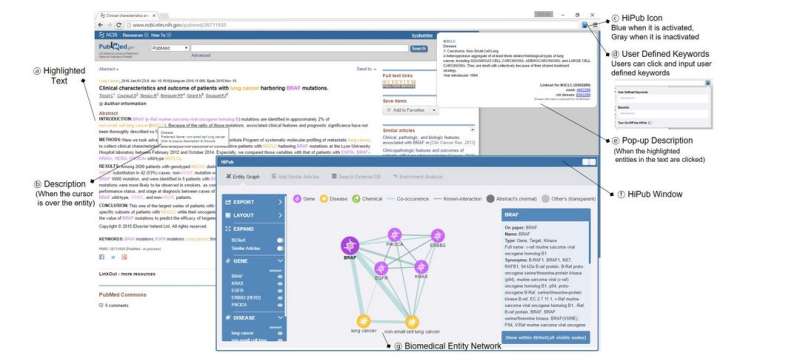
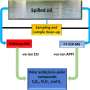
Every day, more than 3,000 new abstracts are uploaded to PubMed, the main biomedical literature reference database. Even in a researcher's narrowly-defined field, it is impossible to stay on top of the ever-evolving webs of interconnections between these papers. For example, a new gene is described - might it be relevant to a researcher's specialty? It could take many painstaking hours of searching to discover the answer. Now a new tool developed in the A.C. Tan lab at the University of Colorado Cancer Center and described today in the journal Bioinformatics helps researchers make these connections. The free tool, HiPub, is available for download as plugin for the Chrome web browser.
"HiPub looks through all this text and tries to recognize what is being called genes, proteins, drugs and diseases. It extracts this information and visualizes it in a network. Especially in molecular biology or cancer biology, it's useful to see the connections between these things in their biological context," says Aik Choon Tan, PhD, investigator at the CU Cancer Center and associate professor at the CU School of Medicine.
Tan gives the example of a hypothetical researcher who reads a paper exploring the genes KRAS and MEK, known to influence the development of certain cancers. "The researcher wants to know if these genes have any relevance to her specialty, maybe something like p53 [another gene known to influence cancer]."
The researcher queries "P53" along with the new article and HiPub visualizes how the researcher's interest is connected to the genes in this new paper. If connections seem compelling, the researcher could design experiments to test these links.
"The idea of text mining isn't new," Tan says. "Computer scientists have been doing it for ten or twenty years. But the real application of text mining in biomedical research is very limited. HiPub is a way to use text mining to streamline the process of knowledge discovery."
More information: Kyubum Lee et al, HiPub: Translating PubMed and PMC Texts to Networks for Knowledge Discovery, Bioinformatics (2016). DOI: 10.1093/bioinformatics/btw511
Journal information: Bioinformatics
Provided by CU Anschutz Medical Campus