Successful recycling: Protein quality control in the cell
A team led by MDC researcher Annika Weber has pinpointed the efficient mechanism used by cells to label faulty proteins. The findings, which provide important insights into the functioning of protein quality control in the cell, have now been published in the journal Molecular Cell.
Proteins perform a wide range of tasks within cells. For everything to run smoothly, the cell must make sure that all the necessary proteins are available and that they are in good condition. Faulty proteins are labeled as such and then recycled. Ligase complexes are able to identify aberrant proteins and attach a small regulatory protein called ubiquitin (Ub) to mark them as faulty. However, further steps are needed before the protein degradation pathway is triggered. Mono-ubiquitination is not enough: a specific chain of multiple Ub molecules must be formed, with the molecules linked to one another at a specific point. Annika Weber and her colleagues from the working group headed by Thomas Sommer at the MDC have now discovered how this chain is formed at the Doa10 ligase complex.
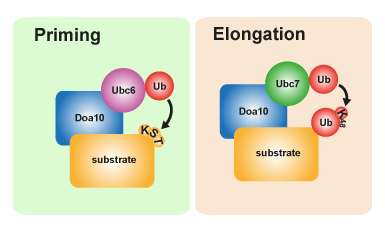
In the priming stage, aberrant proteins are marked with ubiquitin as faulty. To start the degradation process, a second enzyme (Ubc7) is needed to synthesize a ubiquitin chain. Graphics: MDC/Annika Weber.Doa10 plays a key role in protein quality control in the endoplasmic reticulum and the nucleus, monitoring the state of many different proteins in the process. When generating the protein degradation signal, the Ubc6 enzyme first attaches a ubiquitin molecule to the faulty protein. Once this initiation stage (priming) is completed, Ubc7—another enzyme—comes into play, catalyzing the process of chain extension so that a homogenous chain of multiple Ub molecules is formed. As soon as the chain is complete, the targeted protein is sorted to the degradation pathway. Two enzymes are therefore needed to trigger this signal pathway. The research team also discovered that in this special process the primary ubiquitin is not only attached to the amino acid lysine on the defective protein but also to serine residues. These amino acid residues are the "docking points" for the ubiquitin chain on the protein. "This enables faulty proteins to be processed even if no accessible lysine residues are exposed, which makes the degradation process particularly flexible and enables cells to increase the success rate of their protein quality control," says Annika Weber, one of the two lead authors of the paper and a PhD student in Thomas Sommer's Intracellular Proteolysis working group.
A large number of different proteins in a cell have to be degraded—some 30 percent of all cellular protein structures formed by folding of amino acid chains are faulty. The problem for the cells is that these incorrectly folded proteins do not have a uniform structure, making it difficult to identify all of them correctly. If breakdown of these "useless" proteins goes wrong, they are deposited in the cell and disturb its homeostasis. This can lead to death of the cell and trigger a number of diseases, including neurodegenerative disorders such as Alzheimer's and Parkinson's.
More information: Molecular Cell, DOI: 10.1016/j.molcel.2016.07.020
Journal information: Molecular Cell
Provided by Max Delbrück Center for Molecular Medicine in the Helmholtz Association