Scientists disable infectious bacteria by removing key protein
Scientists at the John Innes Centre and the University of East Anglia have made an exciting discovery that could provide a new way to prevent bacterial infections in both humans and plants without triggering multi-drug resistance in bacteria.
When bacteria infect either a plant or a human they first have to move across the surface to a likely site of infection. Without this migration, the bacteria find it difficult to get inside the host and are far less able to cause infection.
The research team, led by Dr Jacob Malone from the JIC and UEA's School of Biological Sciences, was curious about why there were high levels of a particular protein in bacteria when they came into contact with plants. Upon investigating this protein further they discovered it is an important, high-level controller of bacterial movement during plant and human infection. This protein could become a new target against which to develop anti-infective drugs and because the bacteria aren't killed outright, also reduces the likelihood of the bacteria evolving to develop resistance to the drugs.
Dr Malone and his team study a type of bacteria called Pseudomonas, of which there are dozens of different species, including the pathogen P. aeruginosa, which causes around 7% of hospital acquired infections in the UK and is a major cause of mortality in cystic fibrosis patients. Understanding these bacteria is medically very important.

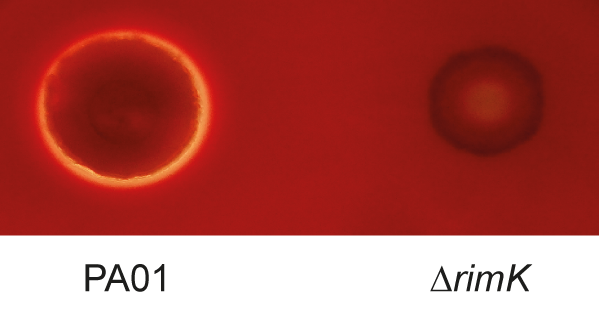
They have found that the ability of Pseudomonas to cause infection is compromised at an early stage when one key protein is removed from the bacterium.
The key protein is called RimK, and is present in hundreds of species of bacteria, including several important human pathogens, but its biological function has remained largely unknown.
Dr Malone's team discovered a completely new function for RimK. When they removed it from bacteria, those bacteria weren't able to move properly, which in turn affected their ability to initiate infections. When scientists examined the effect of RimK deletion on plants they found that spraying a plant with RimK deleted bacteria resulted in milder disease symptoms compared to when wild type bacteria were sprayed on the plant. However, if the early stages of infection were bypassed, for example, by injecting or forcing the bacteria into plant tissue, the RimK mutant bacteria were able to infect normally. This suggests that RimK is important during the early stages of (plant) infection only. Similar results were seen for both plant and human pathogenic Pseudomonas, suggesting a common mechanism for RimK activity in both types of bacteria.
In their paper published in PLoS Genetics, Dr Malone's team showed that RimK works to control the way that many other proteins are made within bacteria. The research suggests that when a bacterium senses that it is in a new place to grow, such as on a plant leaf or on a human cell, it adapts to that environment by changing the production of hundreds of proteins. The RimK protein controls this change, making sure that the bacteria are able to move around and thrive in their new environment. When bacteria are modified in the laboratory so they don't contain a RimK protein, this process doesn't work properly and the bacteria are out of sync with their surroundings, meaning they don't migrate when they should.
Dr Malone said: "We have found a completely new way in which bacteria control their responses to the environment: by modifying their production of proteins. This affects how a bacteria that grows on humans and plants can infect its host - if you disrupt that process, they find it difficult to start an infection."
Dr Malone's team hope that their insight into how the RimK-regulatory process works will influence future medical and plant research, by targeting this protein to help prevent both human and plant diseases.
Dr Malone said: "This information will be extremely useful for researchers working on plant and human diseases. Many people will have heard about the emergence of multidrug resistant bacteria - if we can target a protein that specifically controls infection rather than killing the bacteria outright, the bacteria are less likely to evolve and become resistant. This finding could give scientists a new target against which to develop anti-infective drugs."
More information: 'Adaptive remodelling of the bacterial proteome by specific ribosomal modification regulates Pseudomonas infection and niche colonisation' PLoS Genetics, 2016. dx.doi.org/10.1371/journal.pgen.1005837
Journal information: PLoS Genetics
Provided by John Innes Centre