One, two, four, eight – lessons from dividing cells
The IMCB was established in 1987 at the National University of Singapore before becoming an autonomous research institute of A*STAR and moving to Biopolis in 2004. The IMCB strives to maintain the scientific excellence of principal investigator-driven research and at the same time aims to promote collaborative team-based projects of medical and industrial relevance. Funded primarily by the A*STAR Biomedical Research Council, the IMCB's research activities focus on four major fields: animal models of development and disease, cancer genetics and therapeutics, cell biology in health and disease, and structural biology and drug discovery.
The cell cycle is a highly orchestrated developmental process that leads to the reproduction of a cell. During the cell cycle, faithful replication of a cell's genetic material followed by its accurate segregation ensures that the two daughter cells each inherit an exact copy of the original DNA sequence. Maintaining genomic stability across cell generations relies on the perfect fidelity of these processes. Due to the large sizes of eukaryotic genomes, ranging from 10 million to more than a billion base pairs, accurate and timely replication of DNA is a fundamental challenge during every cell cycle.
As cells progress through the cell cycle, they are exposed to internal and external stresses that may compromise genomic integrity. In humans, genomic instability contributes to a spectrum of pathologies, including cancers, birth defects, developmental abnormalities and a group of diseases collectively known as the chromosome instability syndrome. Given such high stakes, eukaryotic cells have evolved multiple mechanisms to safeguard their genome for cell survival. Mechanisms strictly regulate DNA replication so that each segment of the chromosome is only duplicated once per cell cycle and surveillance mechanisms known as checkpoint controls respond to cues of replicative stress or damaged DNA, transiently halting cell cycle progression until the damage is repaired.
I work in the areas of DNA damage and cell cycle regulation. The cell cycle is like an intricately choreographed ballet involving a repertoire of dancers—various protein complexes—to take the performance through its different acts. Right on cue, all dancers need to be precise in their timing as they enter the stage and in execution as they waltz and pirouette. When someone trips up, there is a pause while the offending event is sorted out. Only then does the choreography pick up to culminate in a grand finale, be it a newly budded yeast cell or one of the many new cells in a little frog embryo. Just imagine, the fertilized egg of the South African clawed toad, Xenopus laevis, undergoes 12 synchronous rounds of cell division in about 8 hours, sub dividing the large single-celled egg into approximately 4,000 smaller cells. Perfect execution, every time!
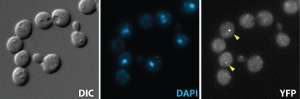
My graduate training was in the area of DNA replication using a cell-free system derived from Xenopus eggs and also using mouse primary cells. For my post-doctoral work, I use the budding yeast, Saccharomyces cerevisiae, as a model system to study the DNA damage response. The availability of genetic tools and its relative ease of manipulation make S. cerevisiae a suitable model system to tackle questions at the molecular level. I now study a phenomenon termed adaptation, which is when cells resume progress through the cell cycle in the presence of irreparable lesions. This means they then undergo cell division with damaged DNA. Adaptation was first described in S. cerevisiae, and has since been observed in other eukaryotic systems.
Adaptation is an interesting cellular response when we consider the significant resources that cells employ to safeguard genomic stability. During normal DNA damage response, cells can either resume the cell cycle when the damage has been successfully repaired, or undergo programmed cell-death/become senescent should the lesion be irreparable. Aberrant DNA content is thus not passed on to future generations. On the other hand, cell division in the presence of unrepaired DNA lesions can lead to accumulation of chromosomal aberrations, which in multicellular organisms may play a role in cancer development. The loss of checkpoint function is widely thought to be an early event in tumorigenesis. By probing the mechanism(s) that leads to cells extinguishing checkpoint signaling in the presence of the DNA damage, we hope to gain a fundamental understanding of an event that possibly confers survival advantages to cancer cells types. And that would be half the battle in thinking about how to treat this disease.
So give up your secrets, budding yeast!