New nanoscopic tools to study ligand-binding of receptors and quantifying two ligand-binding sites
Signalling processes in organisms are governed by specific extracellular and intracellular interactions and involve hundreds of different functionally highly versatile receptors situated in cell membranes. For scientists wishing to understand signalling processes the situation is made more complex by the receptors not only being unevenly distributed and often able to bind more than one ligand but also by the same type of receptor being able to bind a ligand strongly, weakly or not all. New methods that allow precise quantifications of such complex interactions are urgently required.
A new high resolution method developed by an international team of scientists including Robert Tampé and Ralph Wieneke from Goethe University Frankfurt now allows for the first time precise identification and quantification of interactions of a receptor with two ligands simultaneously. The new method has been published in the latest edition of the journal Nature Communications.
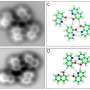
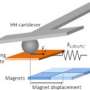
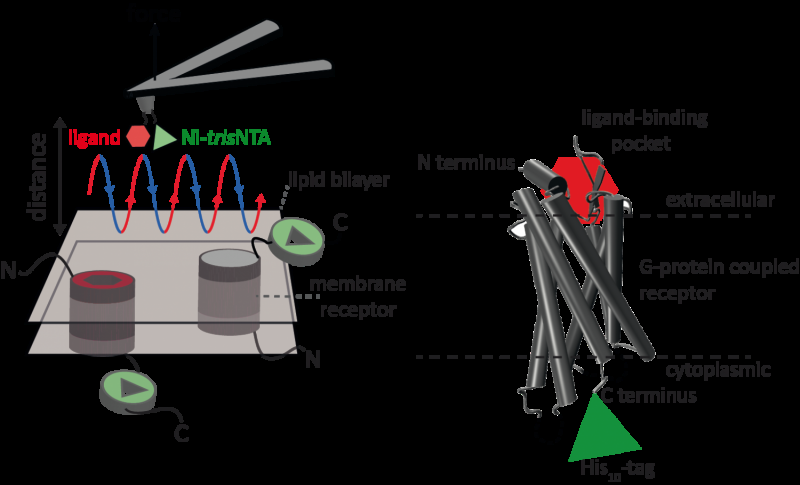
Atomic force microscopy (AFM) is a powerful technique for nanoscale characterization of surfaces. It makes use of a cantilever with an extremely fine tip. Force-distance curve-based atomic force microscopy (FD-based AFM) combines high-resolution imaging and single-molecule force spectroscopy. In studies using biological samples, the AFM tip approaches and retracts from the sample for each pixel. FD-based AFM methods use different coatings of the AFM tip as a toolbox and such methods have made impressive progress in recent years. For the detection of specific binding sites FD-based AFM requires tethering of a ligand to the AFM tip. While contouring protein complexes in a membrane such functionalized AFM tips can then measure the interactions of the tethered ligand to the protein. It had not been possible to image single membrane receptors and simultaneously detect their interactions with more than one ligand, but the new method has overcome this hurdle.
For their proof of principle the scientists used the human protease-activated receptor 1 (PAR1), one of the large family of G-protein-coupled membrane receptors. GPCRs mediate most cellular responses to hormones and neurotransmitters, as well as being responsible for vision, olfaction and taste. GPCRs can coexist in different functional states in the cell membrane and can bind various ligands at different strength or affinity. The GPCR PAR1 is activated by the coagulation protease thrombin which triggers signalling cascades to initiate cellular responses that help orchestrate haemostasis, thrombosis, inflammation and possibly also tissue repair. With the aid of their new FD-based AFM method human PAR1 in proteoliposomes could be imaged while simultaneously detecting extracellular and intracellular interactions of PAR1 with two ligands. The surface chemistry and nanoscopic method developed are applicable to a range of biological systems in vitro and in vivo.
More information: Moritz Pfreundschuh et al. Identifying and quantifying two ligand-binding sites while imaging native human membrane receptors by AFM, Nature Communications (2015). DOI: 10.1038/ncomms9857
Journal information: Nature Communications
Provided by Goethe University Frankfurt