World's fastest algorithm for recognising regular DNA sequences
A mathematical algorithm jointly developed by EURAC and the University of Bolzano (unibz) now permits exceptionally rapid recognition of regular DNA sequences. The previous time of 20 days is reduced to just 5 hours under the new method. Its efficiency and methodological rigour has now led to the algorithm's incorporation in the world's most widely-used DNA-analysis software. This momentous scientific breakthrough is the work of Daniel Taliun. Today at the faculty of Computer Science of the Free University of Bolzano, he discussed his doctoral thesis in information technology, completed at the EURAC Center for Biomedicine.
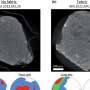
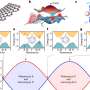
DNA is made up of 3 billion bases, or letters, with the sequence formed of stable segments interspersed with breakpoints. Stable segments are inherited as a single block, while the breaks allow successive sequences to recombine in new ways, ensuring genetic variation between people. The rapid recognition of regular sequences is of great value, as it allows for much more straightforward representation of DNA and for greater precision and speed in identifying those areas of DNA associated with disease. The method developed by Daniel Taliun at the EURAC's Center for Biomedicine and the University of Bolzano is of great utility; the new algorithm processes the entire DNA sequence in 1 percent of the time previously required, down from 20 days to just 5 hours.
"The results caught the attention of the leaders of PLINK, the most widely used software globally for genetic data analysis, who asked us if they could integrate our algorithm into their program," explains Cristian Pattaro, head of the biostatistics group at EURAC's Center for Biomedicine and the research group's specialist on aspects related to genetics and biostatistics.
"This project combines mathematics with information technology and genetics, and has merged the skill sets of two organisations. The University of Bolzano and EURAC have applied their areas of specialisation to achieve a level of excellence that has seen us both working outside of our usual fields of research," says Johann Gamper, professor at the Faculty of Computer Science of the University of Bolzano and supervisor of Daniel's PhD course.
The new algorithm can be applied both in the analysis of the genetic causes of disease and in population genetics. In disease analysis, the recognition of regular DNA segments allows for greater precision in the search for genetic variations associated with illness, in that it allows the examination to be narrowed down to a smaller segment. In population genetics, on the other hand, the recognition of a succession of regular sequences and breakpoints provides information on the study of background genetics, and the researchers have seen that these successions are relatively stable within a single population but may alter between differing populations.
Daniel Taliun returned to Bolzano from the United States for his doctoral viva. "The results of the research have achieved great resonance internationally, and this has led me to obtain a post as researcher at the Department of Biostatistics of the University of Michigan, one of the world's leading centres," concludes Dr. Taliun who, in developing his algorithm, formulated and demonstrated new mathematical theorems.
Provided by European Academy of Bozen/Bolzano