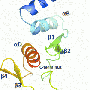3D reconstruction of a vital interaction
Researchers at IBS (CEA/CNRS/Joseph Fourier University) have succeeded for the first time in observing, on an atomic scale, the path taken and the successive changes in form undergone by a disordered vital protein, from its free state to the moment when it binds to another viral protein. The dynamics and mechanisms of this protein interaction, which is involved in the multiplication of the Sendai virus, yield information that could lead to the development of novel anti-viral drugs. This work was published in the Journal of the American Chemical Society on January 20, 2015.
Disordered proteins are very common, both in eukaryotes and in some viruses, and are involved in certain important biological mechanisms and many infectious diseases. The methods used in structural biology today, however, such as crystallography, make no allowance for the flexibility of these proteins, even though their ability to change shape is a key factor in their biological activity.
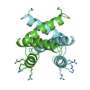
A research team at IBS (CEA/CNRS/Joseph Fourier University) studied the interaction mechanisms between two proteins of the Sendai virus, which is closely related to the human measles virus. The two proteins in question were a nucleoprotein and a phosphoprotein that play a vital role in the multiplication of the virus. Their interaction triggers viral RNA replication, which leads to the infection of the host organism. The researchers used nuclear magnetic resonance (NMR) to reconstruct on the scale of an atom the various stages of the transition from the free, flexible form of the nucleoprotein (NT, shown in yellow below, with its active site in green) to its binding to the surface of the phosphoprotein (PX in orange below) to locate the interaction site.
The active site of the free NT very rapidly adopts different conformations. Only one of them, a helix of a particular length, can interact with PX.
This methodological approach can be used to obtain a 3D reconstruction of many biological mechanisms and, in particular, improve our understanding of how disordered proteins function. More importantly, it could be used to develop drugs that directly target these molecules, which are often involved in human diseases.
Journal information: Journal of the American Chemical Society
Provided by French Alternative Energies and Atomic Energy Commission