RCas9: A programmable RNA editing tool
A powerful scientific tool for editing the DNA instructions in a genome can now also be applied to RNA, the molecule that translates DNA's genetic instructions into the production of proteins. A team of researchers with Berkeley Lab and the University of California (UC) Berkeley has demonstrated a means by which the CRISPR/Cas9 protein complex can be programmed to recognize and cleave RNA at sequence-specific target sites. This finding has the potential to transform the study of RNA function by paving the way for direct RNA transcript detection, analysis and manipulation.
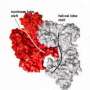
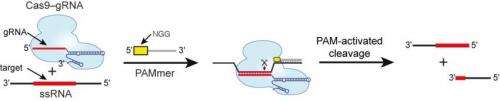
A team led by Jennifer Doudna, biochemist and leading authority on the CRISPR/Cas9 complex, showed how the Cas9 enzyme can work with short DNA sequences known as "PAM," for protospacer adjacent motif, to identify and bind with specific site of single-stranded RNA (ssRNA). They are designating this RNA-targeting CRISPR/Cas9 complex as RCas9.
"Using specially designed PAM-presenting oligonucleotides, or PAMmers, RCas9 can be specifically directed to bind or cut RNA targets while avoiding corresponding DNA sequences, or it can be used to isolate specific endogenous messenger RNA from cells," says Doudna, who holds joint appointments with Berkeley Lab's Physical Biosciences Division and UC Berkeley's Department of Molecular and Cell Biology and Department of Chemistry, and is also an investigator with the Howard Hughes Medical Institute (HHMI). "Our results reveal a fundamental connection between PAM binding and substrate selection by RCas9, and highlight the utility of RCas9 for programmable RNA transcript recognition without the need for genetically introduced tags."
From safer, more effective medicines and clean, green, renewable fuels, to the clean-up and restoration of our air, water and land, the potential is there for genetically engineered bacteria and other microbes to produce valuable goods and perform critical services. To exploit the vast potential of microbes, scientists must be able to precisely edit their genetic information.
In recent years, the CRISPR/Cas complex has emerged as one of the most effective tools for doing this. CRISPR, which stands for Clustered Regularly Interspaced Short Palindromic Repeats, is a central part of the bacterial immune system and handles sequence recognition. Cas9 - Cas stands for CRISPR-assisted - is an RNA-guided enzyme that handles the sniping of DNA strands at the specified sequence site.
Together, CRISPR and Cas9 can be used to precisely edit the DNA instructions in a targeted genome for making desired types of proteins. The DNA is cut at a specific location so that old DNA instructions can be removed and/or new instructions inserted. Until now, it was thought that Cas9 could not be used on the RNA molecules that transcribe those DNA instructions into the desired proteins.
"Just as Cas9 can be used to cut or bind DNA in a sequence-specific manner, RCas9 can cut or bind RNA in a sequence-specific manner," says Mitchell O'Connell, a member of Doudna's research group and the lead author of a paper in Nature that describes this research titled "Programmable RNA recognition and cleavage by CRISPR/Cas9." Doudna is the corresponding author. Other co-authors are Benjamin Oakes, Samuel Sternberg, Alexandra East Seletsky and Matias Kaplan.
In an earlier study, Doudna and her group showed that the genome editing ability of Cas9 is made possible by presence of PAM, which marks where cutting is to commence and activates the enzyme's strand-cleaving activity. In this latest study, Doudna, Mitchell and their collaborators show that PAMmers, in a similar manner, can also stimulate site-specific endonucleolytic cleavage of ssRNA targets. They used Cas9 enzymes from the bacterium Streptococcus pyogenes to perform a variety of in vitro cleavage experiments using a panel of RNA and DNA targets.
"While RNA interference has proven useful for manipulating gene regulation in certain organisms, there has been a strong motivation to develop an orthogonal nucleic-acid-based RNA-recognition system such as RCas9," Doudna says. "The molecular basis for RNA recognition by RCas9 is now clear and requires only the design and synthesis of a matching guide RNA and complementary PAMmer."
The researchers envision a wide range of potential applications for RCas9. For example, an RCas9 tethered to a protein translation initiation factor and targeted to a specific mRNA could essentially act as a designer translation factor to "up-" or "down-" regulate protein synthesis from that mRNA.
"Tethering RCas9 to beads could be used to isolate RNA or native RNA–protein complexes of interest from cells for downstream analysis or assays," Mitchell says. "RCsa9 fused to select protein domains could promote or exclude specific introns or exons, and RCas9 tethered to a fluorescent proteins could be used to observe RNA localization and transport in living cells."
Journal information: Nature
Provided by Lawrence Berkeley National Laboratory