Protein secrets of Ebola virus
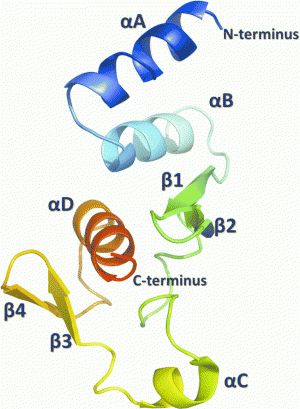
The current Ebola virus outbreak in West Africa, which has claimed more than 2000 lives, has highlighted the need for a deeper understanding of the molecular biology of the virus that could be critical in the development of vaccines or antiviral drugs to treat or prevent Ebola hemorrhagic fever. Now, a team at the University of Virginia (UVA), USA – under the leadership of Dr Dan Engel, a virologist, and Dr Zygmunt Derewenda, a structural biologist – has obtained the crystal structure of a key protein involved in Ebola virus replication, the C-terminal domain of the Zaire Ebola virus nucleoprotein (NP).
The team explains that their structure reveals a novel tertiary fold that is expected to lead to insights into how the viral nucleocapsid is assembled in infected cells. The structure could also provide a basis for the design of drugs to halt infection in humans. "The structure is unique in the RNA virus world," Derewenda explains. "It is not found in viruses that cause influenza, rabies or other diseases." It distantly resembles the β-grasp protein motif found in ubiquitin, most likely the result of convergent evolution.
Like many other related viruses, Ebola virus contains a negative-sense, single-stranded RNA that encodes seven different proteins, one of which is known as the nucleoprotein (NP) for its ability to interact with the viral RNA genome. It is the most abundant viral protein found in infected cells and also inside the viral nucleocapsid. While five of the seven viral proteins have succumbed to structural characterization by X-ray crystallography, NP so far has resisted such attempts, although analogous proteins from other viruses have had their structures analysed.
The UVA team produced the Ebola protein using an engineered form of Escherichia coli bacteria as a protein factory. This allowed them to identify the boundaries of two globular domains and to crystallize the unique C-terminal domain spanning amino-acid residues 641 to 739. The study revealed a molecular architecture unseen so far among known proteins, the team says. There is existing evidence that the newly characterized domain is involved in transcription and the self-assembly of the viral nucleocapsid. As such, the results obtained by the UVA team will be useful in deciphering precisely how these various functions are accomplished by the virus; such a detailed description offers up a potential target for the design of anti-viral drugs.
More information: Dziubanska et al. (2014). Acta Cryst. D70, 2420-2429; DOI: 10.1107/S1399004714014710
Provided by International Union of Crystallography