First metatranscriptome of bee gut finds 19 different bacterial phyla

The digestive tract of the world's most important agricultural pollinator, the honey bee, is a complex fermenting tank that serves up energy-providing short-chain fatty acids thanks to a host of microbial groups that reside in what an Indiana University biologist has described as an intensely intertwined and entangled microbiome.
Work from the laboratory of Indiana University biologist Irene Newton to create the first metatranscriptome of the honey bee gut has shed new light on how those organisms work together and apart from one another to carry out the dominant function of the gut microbiome: carbohydrate metabolism.
"The honey bee eats a primarily plant-based diet made up of foraged foods such as nectar and pollen," said Newton, an assistant professor of biology in the College of Arts and Sciences' Department of Biology at IU Bloomington. "Therefore, we focused our analysis on processes relating to carbohydrate metabolism, which are most relevant to the honey bee and which also dominated our transcriptomic dataset.
"In the honey bee gut microbiome, we saw expression of genes involved in fermentative processes, including the production of short-chain fatty acids, known to serve as an energy source and directly modulate the immune response of other animal hosts."
Genes matching 19 bacterial phyla were identified, but dominating the communities were bacterial groups representing bacilli, gamma-proteobacteria and actinobacteria. Looking at how the entire group of microbiota could metabolize carbon-rich food sources by taking up sugars and fermenting carbohydrates, the team then developed a model of metabolism for the various microbial members.
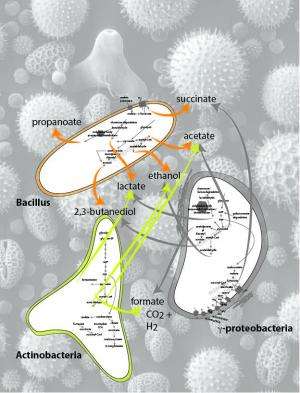
One prominent microbial class in the bee gut, bacilli, is not able to make amino acids, but the other prominent classes—actinobacteria and gamma-proteobacteria—are believed to be able to synthesize all essential amino acids and may provide them to other members of the microbial community and to the bees themselves.
Bacilli, on the other hand, were found to encode and express enzymes involved in the production of short-chain fatty acids, such as acetate and lactate, that may be used by the host bee and by other microbial community members. As an interesting side note, two potential products of metabolism by the bacilli include a carbon storage and anti-freeze compound called 2,3-butanediol, which is used by microbes to prevent intracellular acidification, and acetoin, a chemical that produces a butter flavor in fermented foods.
The team's sequencing strategy also uncovered historically overlooked bacterial groups in their dataset like clostridia, enterobacteriaceae and flavobacteriaceae. Clostridia, which had never been considered a significant contributor to the honey bee microbiome, contributed 46 different genes in the metatranscriptome—a study of the complete set of messenger RNA molecules produced in the cells of the entire group of interacting organisms—including genes involved in metabolism of short-chain fatty acids.
Finally, the team tested some of the predictions based on metatranscriptomic data using community metabolic profiling, an assay that allowed the researchers to detect the ability of a microbial community to utilize specific carbon compounds. The team found that the bee gut community was able to utilize amino acids as well as a large array of saccharides and organic acids, supporting their predictions based on metatranscriptomics. The researchers also found differences between individual bees with regard to both metatranscriptome composition and metabolic profiling, suggesting that the bee microbiome composition and function can differ dramatically from bee to bee.
"We saw major differences in predicted metabolic pathways and in carbon utilization between individual bees from the same hive," Newton said of findings published in the journal Environmental Microbiology. "This result suggests that key environmental or life history variables like age, genetics, diet and season can dramatically affect gut microbiome composition. Understanding the mechanism behind these community shifts will be critical for understanding how gut microbiome composition and activity relate to overall honey bee health and nutrition."
Journal information: Environmental Microbiology
Provided by Indiana University


















