Deeper insights into protein folding
Investigating the structure and dynamics of so-called Meso-Bio-Nano (MBN) systems—micron-sized biological or nanotechnology entities—is a rapidly expanding field of science. Now, scientists Alexander Yakubovich and Andrey Solov'yov from MBN Research Centre in Frankfurt, Germany, have produced a new theoretical study of a protein macromolecule changing from a coil structural conformation to a globular one. Their statistic mechanics model, just published in the European Physical Journal D, describes the thermodynamic properties of real proteins in an aqueous environment, using a minimal number of free physical parameters.
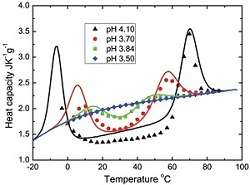
In this work, the authors confirmed the validity of their theoretical calculation of dependencies of the protein heat capacities on temperature by comparing it with the corresponding experimental measurements for two proteins, namely an enzyme called staphylococcal nuclease and an oxygen and iron carrier protein called metmyoglobin. Sudden changes in temperature could result in the loss of a protein's three-dimensional structure and function. Thus, these findings could contribute to our understanding of high-energy ions therapy on biological cells.
In this work, the authors focus on the folding and unfolding of globular proteins at various levels of temperature in an aqueous environment. Their statistical mechanics model is inspired by a pre-existing model of solvation of hydrophobic hydrocarbons. This leads to establishing the so-called partition function of this globular protein in water environment. In turn, this helps to determine all of the protein's thermodynamic characteristics at equilibrium. These include its heat capacity and the average number of amino acids in an unfolded conformation.
The study validates the use of an approximation of three stages of macromolecular complexes undergoing folding and unfolding transformations, instead of using fitting parameters as previously done. These results also significantly expand the possibilities of quantitative description of the structure conformation processes for other proteins obeying simple folding kinetics and complex multi-domain proteins with peculiar folding profiles.
More information: A. V. Yakubovich and A. V. Solov'yov (2014), Quantitative thermodynamic model for globular protein folding, European Physical Journal D, DOI: 10.1140/epjd/e2014-50097-3
Journal information: European Physical Journal D
Provided by Springer