Enzyme 'wrench' could be key to stronger, more effective antibiotics
Builders and factory workers know that getting a job done right requires precision and specialized tools. The same is true when you're building antibiotic compounds at the molecular level. New findings from North Carolina State University may turn an enzyme that acts as a specialized "wrench" in antibiotic assembly into a set of wrenches that will allow for greater customization. By modifying this enzyme, scientists hope to be able to design and synthesize stronger, more adaptable antibiotics from less expensive, natural compounds.
Kirromycin is a commonly known antibiotic that can be created through natural synthesis; that is, it doesn't have to be made in a chemistry lab. Nature creates compounds like kirromycin through a factory-like assembly line of enzymes where each performs a specific function, snapping different fragments of molecules together like a jigsaw puzzle. Understanding this process on the molecular level could give chemists the ability to piggyback on nature, synthesizing new antibiotics and cancer drugs with less waste and expense.
NC State chemist Gavin Williams looked at one enzyme in the kirromycin assembly line – KirCII – which is responsible for installing a molecular fragment of kirromycin at one key location. "KirCII is a linchpin enzyme in the assembly," Williams says. "Natural compounds like kirromycin get built in pieces, with small modules, or blocks of enzymes, linking up sections of the compound in a certain order. Enzymes like KirCII are the wrenches that install the molecular pieces – without them, the molecule doesn't finish assembling properly."
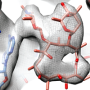
Williams and his team performed a molecular analysis of KirCII to determine why and how it latches onto a specific protein within the kirromycin assembly line. They saw that the enzyme has electrical charges on its surface that are complimentary to opposite charges on the surface of the protein it binds with. When KirCII finds that protein, the charges match up and it snaps into place.
"We were able to see which areas on KirCII had charges that worked with the target protein," Williams says. "Hopefully we will be able to use this information to introduce complimentary charges onto the surface of other proteins we want KirCII to bind with.
"Right now KirCII is just one wrench. By modifying it to fit other proteins, we could turn it into a set of different wrenches and create totally different antibiotics. Kirromycin isn't very useful right now, but by using KirCII to install pieces from other antibiotics, we'll be able to mix and match and create new, stronger antibiotics."
More information: "Reprogramming Acyl Carrier Protein Interactions of an Acyl-CoA Promiscuous trans-Acyltransferase", Zhixia Ye, et al. April 10, 2014 in Chemistry and Biology
Abstract
Protein interactions between acyl carrier proteins (ACPs) and trans-acting acyltransferase domains (trans-ATs) are critical for regioselective extender unit installation by many polyketide synthases, yet little is known regarding the specificity of these interactions, particularly for trans-ATs with unusual extender unit specificities. Currently, the best-studied trans-AT with nonmalonyl specificity is KirCII from kirromycin biosynthesis. Here, we developed an assay to probe ACP interactions based on leveraging the extender unit promiscuity of KirCII. The assay allows us to identify residues on the ACP surface that contribute to specific recognition by KirCII. This information proved sufficient to modify a noncognate ACP from a different biosynthetic system to be a substrate for KirCII. The findings form a foundation for further understanding the specificity of trans-AT:ACP protein interactions and for engineering modular polyketide synthases to produce analogs.
Journal information: Cell Chemical Biology
Provided by North Carolina State University