Functional diversity in bacterial defense mechanism against viral invasion
Bacteria may lack a true immune system, but this does not leave them defenseless against bacteriophage viruses and other pathogens. A system of genomic sequence elements called clustered regularly interspaced short palindromic repeats (CRISPR) and various CRISPR-associated proteins (Cas) help to recognize and destroy foreign genetic material delivered by such invaders.
An international research group led by Akeo Shinkai from the RIKEN SPring-8 Center and John van der Oost of Wageningen University in the Netherlands has now dissected one such CRISPR-Cas pathway, revealing functional insights that also highlight important differences in how these systems operate across bacterial species.
The researchers focused their attention on Thermus thermophilus, a bacterium that thrives at high temperatures and features a relatively simple and compact genome, making it amenable to experimental work. Of the bacterium's multiple CRISPR-Cas pathways, the researchers explored the pathway known as subtype III-B, which targets foreign RNA rather than DNA.
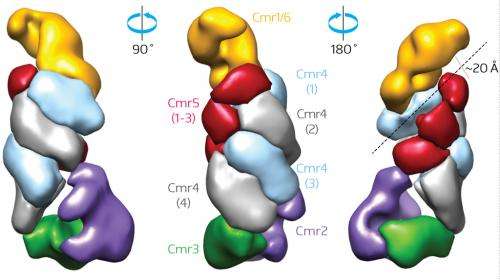
Every CRISPR element contains multiple repeats of gene sequences separated by unique spacer sequences, each of which corresponds to a potential CRISPR-Cas target. Importantly, the CRISPR-Cas system also collects 'trophies' from novel invaders, incorporating their sequence information into new CRISPR-spacer elements, thus enabling future recognition of the same pathogen. These CRISPR genes are transcribed to produce spacer-specific CRISPR RNAs (crRNAs), which combine with a collection of Cas proteins known as the Cmr complex. Shinkai and van der Oost were able to isolate the Cmr complex and identified an elongated structure that they describe as resembling a 'sea worm', with a channel that could potentially accommodate the crRNA strand (Fig. 1).
The researchers were also able to isolate these associated crRNAs and determined that Cmr predominantly uses spacer sequences from just 4 of the 11 CRISPR loci in the T. thermophilus genome. They also identified a surprising mechanism for Cmr-induced cleavage, where the complex cuts at multiple sites at fixed distances along the target, as opposed to the sequence-specific or single-site cleavage mechanisms identified in other CRISPR-Cas pathways. "The T. thermophilus Cmr complex may have evolved to kill bacteriophages as quickly as possible," says Shinkai. "This demonstrates the diversity of the CRISPR-Cas system."
Shinkai and his colleagues now hope to probe the three-dimensional structure of the complex in more depth and to embark on similar analyses for the other CRISPR-Cas pathways active in T. thermophilus. "This will hopefully lead to a more systematic understanding of these systems within the cell and of the diversity of these systems in the microbial world," says Shinkai.
More information: Staals, R. H. J., Agari, Y., Maki-Yonekura, S., Zhu, Y., Taylor, D. W., van Duijn, E., Barendregt, A., Vlot, M., Koehorst, J. J., Sakamoto, K. et al. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Molecular Cell 52, 135–145 (2013). dx.doi.org/10.1016/j.molcel.2013.09.013
Journal information: Molecular Cell
Provided by RIKEN