Improved decoding of DNA for custom medical treatments
One day, doctors will be able to create custom medical treatment plans based on a patient's DNA, pinpointing the root of a patient's illness and making sure treatment will not cause a fatal allergic reaction. Thanks to Technion Professor Amit Meller fantasy is one step closer to being a reality
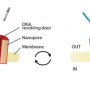
The key to bringing about this revolutionary DNA-based medicine is the quick and accurate decoding of a patient's genome. A genome is the unique sequence of special molecules along a chain of DNA that tells a cell's machinery which proteins to produce, and when. Those crucial genome molecules are called "nucleobases," and are known as adenine, thymine, cytosine, and guanine (or A, T, C, and G, for short). Prof. Meller and his team developed a way to record the As, Ts, Cs, and Gs in a person's DNA by forcing a DNA molecule to slip through a tiny hole – called a "nanopore" – in a tiny silicon chip the size of the head of a nail.
(Just how small is a nanopore? It measures anywhere between 2 and 5 nanometers, or billionths of a meter, in diameter. In comparison, a human hair measures 100 micrometers, or millionths of a meter, in diameter.)
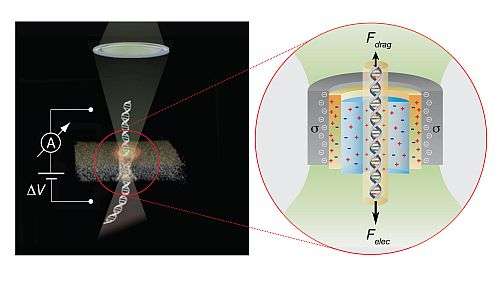
The scientists begin by dunking the DNA molecules in a combination of water and electrically charged salt molecules. As the saltwater flows through the nanopore, it creates an electric current. When a DNA molecule passes through the pore, however, the current is disrupted. And, the amount of current disruption depends on which A, T, C, or G is in the pore.
Therefore, to read the sequence of nucleobases, a scientist simply has to find out how much each base disrupts the electric current. With that information, he could read the sequence of DNA bases simply by logging the sequence of electrical disruptions as a DNA molecule passed through. There's a catch, though. "To do this, each base must stay in the pore long enough to make it very clear how much current it blocks, so that one can correctly identify the nucleobase," says Prof. Meller.
But DNA usually moves too quickly through the nanopores for Meller and his team to decode it. To slow the DNA down, they shone a green laser – no stronger than laser pointers used in classrooms – at the pore, which gave it a negative electric charge. The nanopore then attracted the positively charged potassium atoms in the saltwater. Those atoms, along with some of the water, moved towards the pore, creating a flow that blocked the movement of the DNA. "So, that creates a drag force on the DNA, slowing it down so that each base sites in the nanopore longer," says Prof. Meller.
This method of reading DNA sequences is still under laboratory development. But Meller envisions a future in which the nanopore chip could be built into a portable device—about the size of a smartphone—that could be brought right to the patient.
The Technion research team collaborated with colleagues at Boston University on this project. The team's results were published on the November 3 online edition of Nature Nanotechnology.
Journal information: Nature Nanotechnology
Provided by American Technion Society