Computational approach reveals myriad of protein pairs that combine to regulate gene activity
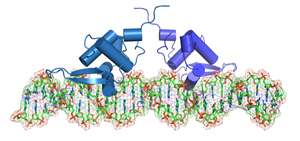
Transcription factor (TF) proteins switch particular genes on and off by homing in on specific sequences of nucleotides scattered throughout the genome. In some cases, this regulation is a team effort, where pairs of TFs—or 'dimers'—come together to modulate one target gene.
A team led by Shyam Prabhakar of the A*STAR Genome Institute of Singapore and Jerzy Tiuryn at the University of Warsaw, Poland, has now devised a computational strategy for mapping TF collaboration at an unprecedented scale. Most TF dimers have been discovered through experiments that focused exclusively on individual TFs or TF pairs. As an alternative, Prabhakar and co-workers identified such binding events on a large scale based on a few basic assumptions.
"We predicted that these dimers would be reasonably compact, with binding sites spaced less than 50 bases apart," he explains, "and that most dimers would have a favorite spacing."
Access was another important consideration; chromosomal DNA is wound around protein cores to form a material known as chromatin, which can be dense enough to thwart TF binding. Prabhakar and co-workers used chromatin maps to flag regions that were 'open' for protein binding. As chromatin structure can vary considerably between cell types, this approach also gave the team an edge in characterizing potential tissue-specific activity of TF dimers.
The researchers used their algorithm to search for combinations of 964 different TF binding sites within regions of open chromatin in 41 different human cell types. Their data predicted 603 potential dimers, including 19 of the 25 pairs that have been identified experimentally to date and many more that were previously unknown. Altogether, these predicted dimers bound almost half a million distinct sites in the genome, vastly exceeding prior predictions.
"I don't think anyone appreciated just how many TF dimers there were in human cells—they were usually treated as an exotic, special case," says Prabhakar. The data also supported his prediction that TFs form fairly rigid complexes with DNA binding sites positioned at a relatively fixed distance from each other, rather than the more flexible mode of dimer interaction supported by some other scientists.
Importantly, Prabhakar and team identified many TF dimers with apparent cell-specific activity, including certain combinations that seem particularly active in tumor cells. For example, various dimers containing the TF FOXA1 were prominently over-represented in prostate cancer cells (see image). Prabhakar and Tiuryn intend to further explore the clinical significance of these and other findings in future experiments with collaborator Ralf Jauch.
More information: Jankowski, A., Szczurek, E., Jauch, R., Tiuryn, J. & Prabhakar, S. Comprehensive prediction in 78 human cell lines reveals rigidity and compactness of transcription factor dimers. Genome Research 23, 1307–1318 (2013). dx.doi.org/10.1101/gr.154922.113
Journal information: Genome Research